Dysbiotic signatures and diagnostic potential of gut microbial markers for inflammatory bowel disease in Korean population
- PMID: 39390011
- PMCID: PMC11467411
- DOI: 10.1038/s41598-024-74002-6
Dysbiotic signatures and diagnostic potential of gut microbial markers for inflammatory bowel disease in Korean population
Abstract
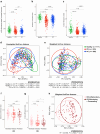
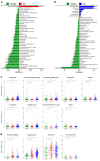
Fecal samples were collected from 640 individuals in Korea, including 523 patients with IBD (223 with Crohn's disease [CD] and 300 with ulcerative colitis [UC]) and 117 healthy controls. The samples were subjected to cross-sectional gut metagenomic analysis using 16 S rRNA sequencing and bioinformatics analysis. Patients with IBD, particularly those with CD, exhibited significantly lower alpha diversities than the healthy subjects. Differential abundance analysis revealed dysbiotic signatures, characterized by an expansion of the genus Escherichia-Shigella in patients with CD. Functional annotations showed that functional pathways related to bacterial pathogenesis and production of hydrogen sulfide (H2S) were strongly upregulated in patients with CD. A dysbiosis score, calculated based on functional characteristics, highly correlated with disease severity. Markers distinguishing between healthy subjects and patients with IBD showed accurate classification based on a small number of microbial taxa, which may be used to diagnose ambiguous cases. These findings confirm the taxonomic and functional dysbiosis of the gut microbiota in patients with IBD, especially those with CD. Taxa indicative of dysbiosis may have significant implications for future clinical research on the management and diagnosis of IBD.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Yilmaz, B. et al. Microbial network disturbances in relapsing refractory Crohn’s disease. Nat. Med.25, 323 (2019). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

