Protein biomarker signatures of preeclampsia - a longitudinal 5000-multiplex proteomics study
- PMID: 39390022
- PMCID: PMC11467422
- DOI: 10.1038/s41598-024-73796-9
Protein biomarker signatures of preeclampsia - a longitudinal 5000-multiplex proteomics study
Abstract
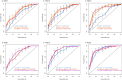
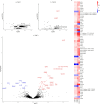
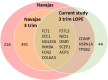
We aimed to explore novel biomarker candidates and biomarker signatures of late-onset preeclampsia (LOPE) by profiling samples collected in a longitudinal discovery cohort with a high-throughput proteomics platform. Using the Somalogic 5000-plex platform, we analyzed proteins in plasma samples collected at three visits (gestational weeks (GW) 12-19, 20-26 and 28-34 in 35 women with LOPE (birth ≥ 34 GW) and 70 healthy pregnant women). To identify biomarker signatures, we combined Elastic Net with Stability Selection for stable variable selection and validated their predictive performance in a validation cohort. The biomarker signature with the highest predictive performance (AUC 0.88 (95% CI 0.85-0.97)) was identified in the last trimester of pregnancy (GW 28-34) and included the Fatty acid amid hydrolase 2 (FAAH2), HtrA serine peptidase 1 (HTRA1) and Interleukin-17 receptor C (IL17RC) together with sFLT1 and maternal age, BMI and nulliparity. Our biomarker signature showed increased or similar predictive performance to the sFLT1/PGF-ratio within our data set, and we were able to validate the biomarker signature in a validation cohort (AUC ≥ 0.90). Further validation of these candidates should be performed using another protein quantification platform in an independent cohort where the negative and positive predictive values can be validly calculated.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Lisonkova, S. & Joseph, K. S. Incidence of preeclampsia: risk factors and outcomes associated with early- versus late-onset disease. Am J Obstet Gynecol 209, 544 e541-544 e512, doi: (2013). 10.1016/j.ajog.2013.08.019 - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

