KLF4 regulates trophoblast function and associates with unexplained recurrent spontaneous abortion
- PMID: 39390495
- PMCID: PMC11465507
- DOI: 10.1186/s12967-024-05707-5
KLF4 regulates trophoblast function and associates with unexplained recurrent spontaneous abortion
Abstract
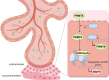
Background: Recurrent spontaneous abortion (RSA) is defined as two or more consecutive spontaneous abortions before 20 weeks with the same spouse [1]. However, approximately 50% of RSA cases of unknown cause are classified as unexplained recurrent spontaneous abortion (URSA). Potential factors include decreased trophoblast cell migration and invasion, leading to impaired placental implantation and maintenance of the normal maternal-fetal interface. However, the mechanism of this pathogenesis remains unknown. In this study, we investigated the potential role and mechanism of KLF4 in regulating URSA by influencing the invasion and migration ability of trophoblast cells.
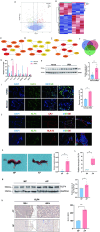
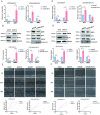
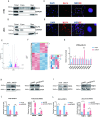
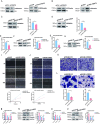
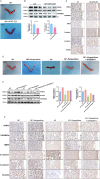
Methods: We firstly identified 817 differentially expressed genes by performing a difference analysis of the dataset GSE121950 [2] related to recurrent abortion, and intersected the top 10 genes obtained respectively by the three algorithms: DMNC, MNC, and EPC using Venn Diagram.To detect the expression levels of core genes, villi samples were obtained from normal pregnant women and patients with URSA. RT-qPCR analysis revealed a significant difference in KLF4 mRNA expression and KLF4 was then analyzed. Trophoblast cell lines HTR8 and JEG3 were used to investigate the effect of KLF4 on trophoblastic function. Wound healing and transwell assays was performed to detect the invasion and migration of trophoblast cells. The expression of epithelial-mesenchymal transition(EMT) molecules were detected by RT-qPCR and western blot. Promoter detection and epigenetic modification were detected by chromatin immunoprecipitation (ChIP) assay. Molecular nuclear localization was detected by immunofluorescence and subcellular fractionation. Miscarried mice model was used to study the effects of KLF4 on URSA induced by reduced trophoblast invasion and migration.
Results: KLF4 is highly expressed in the villi of patients with URSA. KLF4 inhibits the expression level of H3R2ME2a in trophoblast cells by regulating the transcriptional level and nuclear translocation of PRMT6, thereby inhibiting the possible regulatory mechanism of trophoblastic invasion and providing a potential treatment strategy for URSA in vivo.
Conclusions: The KLF4/PRMT6/H3R2ME2a axis regulates mechanisms associated with unexplained recurrent spontaneous abortion by regulating trophoblast function.
Keywords: H3R2ME2a; KLF4; PRMT6; Trophoblast invasion; Unexplained recurrent spontaneous abortion.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures







References
-
- Rai R, Regan L. Recurrent miscarriage. Lancet. 2006;368(9535):601–11. - PubMed
-
- La XL, Wang WJ, Zhang M, Liang L. Definition and multiple factors of recurrent spontaneous abortion. Environ Female Reproductive Health. 2021;1300:231–57. - PubMed
-
- Ibrahaim Y, Hotaling J. Sperm epigenetics and its impact on male fertility, pregnancy loss, and somatic health of future offspring. Sem Reprod Med. 2018;36(3–4):233–5. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

