Dual-ligand PROTACS mediate superior target protein degradation in vitro and therapeutic efficacy in vivo
- PMID: 39391379
- PMCID: PMC11462456
- DOI: 10.1039/d4sc03555k
Dual-ligand PROTACS mediate superior target protein degradation in vitro and therapeutic efficacy in vivo
Abstract
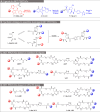
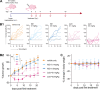
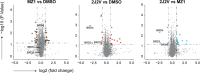
Proteolysis targeting chimeras (PROTACs) are revolutionizing the drug development landscape due to their unique ability to selectively degrade disease-associated proteins. Conventional PROTACs are bivalent entities that induce ubiquitination and subsequent proteolysis of a chosen protein of interest (POI) by forming a ternary complex with an E3 ligase. We hypothesized that dual-ligand PROTACs, featuring two copies each of a POI ligand and an E3 ligase ligand, would facilitate the formation of high-avidity, long-lived ternary complexes inside cells, thereby increasing POI degradation potency. To this end, we developed a convergent synthesis route, using l-aspartic acid as a building block for homodimer synthesis, followed by copper-catalyzed azide-alkyne cycloaddition (CuAAC) to conjugate both dimers through a flexible linker. Dual-ligand PROTACs achieved up to a tenfold increase in degradation efficiency and a hundredfold increase in cytotoxicity in vitro across various cancer cell lines compared to their single-ligand counterparts. Furthermore, dual-ligand PROTACs sustain prolonged protein degradation, up to 60 hours after pulsing and washout. In vivo, in a mouse tumor model, the superior therapeutic activity of dual ligand PROTACs was observed.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
The authors declare no competing financial interest.
Figures




References
-
- Bondeson D. P. Mares A. Smith I. E. Ko E. Campos S. Miah A. H. Mulholland K. E. Routly N. Buckley D. L. Gustafson J. L. Zinn N. Grandi P. Shimamura S. Bergamini G. Faelth-Savitski M. Bantscheff M. Cox C. Gordon D. A. Willard R. R. Flanagan J. J. Casillas L. N. Votta B. J. den Besten W. Famm K. Kruidenier L. Carter P. S. Harling J. D. Churcher I. Crews C. M. Catalytic in vivo protein knockdown by small-molecule PROTACs. Nat. Chem. Biol. 2015;11(8):611–617. doi: 10.1038/nchembio.1858. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

