Germinal center B-cell subgroups in the tumor microenvironment cannot be overlooked: Their involvement in prognosis, immunotherapy response, and treatment resistance in head and neck squamous carcinoma
- PMID: 39391510
- PMCID: PMC11466559
- DOI: 10.1016/j.heliyon.2024.e37726
Germinal center B-cell subgroups in the tumor microenvironment cannot be overlooked: Their involvement in prognosis, immunotherapy response, and treatment resistance in head and neck squamous carcinoma
Abstract
Background: More than 60 % of patients with head and neck squamous carcinoma (HNSCC) are diagnosed at advanced stages and miss radical treatment. This has prompted the need to find new biomarkers to achieve early diagnosis and predict early recurrence and metastasis of tumors.
Methods: Single-cell RNA sequencing (scRNA-seq) data from HNSCC tissues and peripheral blood samples were obtained through the Gene Expression Omnibus (GEO) database (GSE164690) to characterize the B-cell subgroups, differentiation trajectories, and intercellular communication networks in HNSCC and to construct a prognostic model of the associated risks. In addition, this study analyzed the differences in clinical features, immune cell infiltration, functional enrichment, tumor mutational burden (TMB), and drug sensitivity between the high- and low-risk groups.
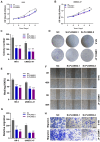
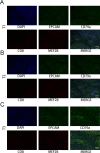
Results: Using scRNA-seq of HNSCC, we classified B and plasma cells into a total of four subgroups: naive B cells (NBs), germinal center B cells (GCBs), memory B cells (MBs), and plasma cells (PCs). Pseudotemporal trajectory analysis revealed that NBs and GCBs were at the early stage of B cell differentiation, while MBs and PCs were at the end. Cellular communication revealed that GCBs acted on tumor cells through the CD99 and SEMA4 signaling pathways. The independent prognostic value, immune cell infiltration, TMB and drug sensitivity assays were validated for the MEF2B+ GCB score groups.
Conclusions: We identified GCBs as B cell-specific prognostic biomarkers for the first time. The MEF2B+ GCB score fills the research gap in the genetic prognostic prediction model of HNSCC and is expected to provide a theoretical basis for finding new therapeutic targets for HNSCC.
Keywords: Drug sensitivity; Germinal center B cells; Head and neck squamous carcinoma; Immune cell infiltration; Single-cell RNA-Seq.
© 2024 The Authors.
Conflict of interest statement
None.
Figures















Similar articles
-
Tumor Mutation Burden, Immune Cell Infiltration, and Construction of Immune-Related Genes Prognostic Model in Head and Neck Cancer.Int J Med Sci. 2021 Jan 1;18(1):226-238. doi: 10.7150/ijms.51064. eCollection 2021. Int J Med Sci. 2021. PMID: 33390791 Free PMC article.
-
The cellular signaling crosstalk between memory B cells and tumor cells in nasopharyngeal carcinoma cannot be overlooked: Their involvement in tumor progression and treatment strategy is significant.J Cancer. 2025 Jan 1;16(1):288-314. doi: 10.7150/jca.101420. eCollection 2025. J Cancer. 2025. PMID: 39744570 Free PMC article.
-
FAT1 as a tumor mutation burden specific gene affects the immunotherapy effect in head and neck squamous cell cancer.Drug Resist Updat. 2024 Sep;76:101095. doi: 10.1016/j.drup.2024.101095. Epub 2024 May 27. Drug Resist Updat. 2024. PMID: 38986165
-
The prognostic value of TMB and the relationship between TMB and immune infiltration in head and neck squamous cell carcinoma: A gene expression-based study.Oral Oncol. 2020 Nov;110:104943. doi: 10.1016/j.oraloncology.2020.104943. Epub 2020 Sep 9. Oral Oncol. 2020. PMID: 32919362
-
FAM3D as a Prognostic Indicator of Head and Neck Squamous Cell Carcinoma Is Associated with Immune Infiltration.Comput Math Methods Med. 2022 Dec 3;2022:5851755. doi: 10.1155/2022/5851755. eCollection 2022. Comput Math Methods Med. 2022. PMID: 36510584 Free PMC article.
Cited by
-
Single-cell insights into HNSCC tumor heterogeneity and programmed cell death pathways.Transl Oncol. 2025 Apr;54:102341. doi: 10.1016/j.tranon.2025.102341. Epub 2025 Mar 10. Transl Oncol. 2025. PMID: 40068384 Free PMC article.
-
Exposing the cellular situation: findings from single cell RNA sequencing in breast cancer.Front Immunol. 2025 Mar 6;16:1539074. doi: 10.3389/fimmu.2025.1539074. eCollection 2025. Front Immunol. 2025. PMID: 40114930 Free PMC article.
-
Single-cell and spatial atlas of glioblastoma heterogeneity: characterizing the PCLAF+ subtype and YEATS4's oncogenic role.Front Immunol. 2025 Jul 25;16:1614549. doi: 10.3389/fimmu.2025.1614549. eCollection 2025. Front Immunol. 2025. PMID: 40787449 Free PMC article.
-
Construction and validation of a chemokine-related gene signature associated with prognosis, clinical significance, and immune microenvironment characteristics in cervical cancer.Discov Oncol. 2025 Jun 15;16(1):1114. doi: 10.1007/s12672-025-02973-7. Discov Oncol. 2025. PMID: 40517358 Free PMC article.
-
MAZ-mediated tumor progression and immune evasion in hormone receptor-positive breast cancer: Targeting tumor microenvironment and PCLAF+ subtype-specific therapy.Transl Oncol. 2025 Feb;52:102280. doi: 10.1016/j.tranon.2025.102280. Epub 2025 Jan 12. Transl Oncol. 2025. PMID: 39805182 Free PMC article.
References
-
- Leemans C.R., Snijders P., Brakenhoff R.H. The molecular landscape of head and neck cancer. Nat. Rev. Cancer. 2018;18:269–282. - PubMed
-
- Carlisle J.W., Steuer C.E., Owonikoko T.K., et al. An update on the immune landscape in lung and head and neck cancers. CA A Cancer J. Clin. 2020;70:505–517. - PubMed
-
- Travassos D.C., Fernandes D., Massucato E., et al. Squamous cell carcinoma antigen as a prognostic marker and its correlation with clinicopathological features in head and neck squamous cell carcinoma: systematic review and meta-analysis. J. Oral Pathol. Med. 2018;47:3–10. - PubMed
-
- Sawant S.S., Zingde S.M., Vaidya M.M. Cytokeratin fragments in the serum: their utility for the management of oral cancer. Oral Oncol. 2008;44:722–732. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

