Harnessing de novo transcriptome sequencing to identify and characterize genes regulating carbohydrate biosynthesis pathways in Salvia guaranitica L
- PMID: 39391775
- PMCID: PMC11464306
- DOI: 10.3389/fpls.2024.1467432
Harnessing de novo transcriptome sequencing to identify and characterize genes regulating carbohydrate biosynthesis pathways in Salvia guaranitica L
Abstract
Introduction: Carbohydrate compounds serve multifaceted roles, from energy sources to stress protectants, found across diverse organisms including bacteria, fungi, and plants. Despite this broad importance, the molecular genetic framework underlying carbohydrate biosynthesis pathways, such as starch, sucrose, and glycolysis/gluconeogenesis in Salvia guaranitica, remains largely unexplored.
Methods: In this study, the Illumina-HiSeq 2500 platform was used to sequence the transcripts of S. guaranitica leaves, generating approximately 8.2 Gb of raw data. After filtering and removing adapter sequences, 38 million reads comprising 210 million high-quality nucleotide bases were obtained. De novo assembly resulted in 75,100 unigenes, which were annotated to establish a comprehensive database for investigating starch, sucrose, and glycolysis biosynthesis. Functional analyses of glucose-6-phosphate isomerase (SgGPI), trehalose-6-phosphate synthase/phosphatase (SgT6PS), and sucrose synthase (SgSUS) were performed using transgenic Arabidopsis thaliana.
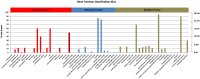
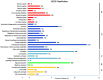
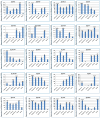
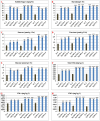
Results: Among the unigenes, 410 were identified as putatively involved in these metabolic pathways, including 175 related to glycolysis/gluconeogenesis and 235 to starch and sucrose biosynthesis. Overexpression of SgGPI, SgT6PS, and SgSUS in transgenic A. thaliana enhanced leaf area, accelerated flower formation, and promoted overall growth compared to wild-type plants.
Discussion: These findings lay a foundation for understanding the roles of starch, sucrose, and glycolysis biosynthesis genes in S. guaranitica, offering insights into future metabolic engineering strategies for enhancing the production of valuable carbohydrate compounds in S. guaranitica or other plants.
Keywords: Salvia guaranitica; functional characterization; gluconeogenesis; glycolysis; starch; sucrose; transcriptome; transgenic Arabidopsis thaliana.
Copyright © 2024 Abbas, Al-Huqail, Abdel Kawy, Abdulhai, Albalawi, AlShaqhaa, Alsubeie, Darwish, Abdelhameed, Soudy, Makki, Aljabri, Al-Sulami, Ali and Zayed.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The reviewer OH declared a past co-authorship with the author MA, and declared a shared affiliation with the author MZ to the handling editor at the time of the review.
Figures



References
-
- Abdelhameed A. A., Ali M., Darwish D. B. E., AlShaqhaa M. A., Selim D. A.-F. H., Nagah A., et al. (2024. a). Induced genetic diversity through mutagenesis in wheat gene pool and significant use of SCoT markers to underpin key agronomic traits. BMC Plant Biol. 24, 673. doi: 10.1186/s12870-024-05345-5 - DOI - PMC - PubMed
-
- Abdelhameed A. A., Eissa M. A., El-kholy R. I., Darwish D. B. E., Abeed A. H. A., Soudy F. A., et al. (2024. b). Molecular cloning and expression analysis of geranyllinalool synthase gene (SgGES) from salvia guaranitica plants. Horticulturae 10, 668. doi: 10.3390/horticulturae10070668 - DOI
-
- Abd El-Wahab M., Toaima W., Hamed E. (2015). Effect of different planting locations in Egypt on salvia fruticosa mill. Plants. Egypt. J. Desert Res. 65, 291–307. doi: 10.21608/ejdr.2015.5955 - DOI
-
- Ahmed M., Shahid A. A., Akhtar S., Latif A., Din S., Fanglu M., et al. (2018). Sucrose synthase genes: a way forward for cotton fiber improvement. Biol. (Bratisl). 73, 703–713. doi: 10.2478/s11756-018-0078-6 - DOI
LinkOut - more resources
Full Text Sources

