Metabolic dysfunction-associated steatotic liver disease exhibits sex-specific microbial heterogeneity within intestinal compartments
- PMID: 39391907
- PMCID: PMC11791572
- DOI: 10.3350/cmh.2024.0359
Metabolic dysfunction-associated steatotic liver disease exhibits sex-specific microbial heterogeneity within intestinal compartments
Abstract
Background/aims: Evidence suggests that the gastrointestinal microbiome plays a significant role in the biology of metabolic dysfunction-associated steatotic liver disease (MASLD). However, it remains unclear whether disparities in the gut microbiome across intestinal tissular compartments between the sexes lead to MASLD pathogenesis.
Methods: Sex-specific analyses of microbiome composition in two anatomically distinct regions of the gut, the small intestine and colon, were performed using an experimental model of MASLD. The study involved male and female spontaneously hypertensive rats and the Wistar-Kyoto control rat strain, which were fed either a standard chow diet or a high-fat diet for 12 weeks to induce MASLD (12 rats per group). High-throughput 16S sequencing was used for microbiome analysis.
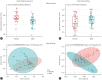
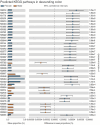
Results: There were significant differences in the overall microbiome composition of male and female rats with MASLD, including variations in topographical gut regions. The beta diversity of the jejunal and colon microbiomes was higher in female rats than in male rats (PERMANOVA p-value=0.001). Sex-specific analysis and discriminant features using LEfSe showed considerable variation in bacterial abundance, along with distinct functional properties, in the jejunum and colon of animals with MASLD. Significantly elevated levels of lipopolysaccharide and protein expression of Toll-like receptor 4 were observed in the livers of male rats with MASLD compared with their female counterparts.
Conclusion: This study uncovered sexual dimorphism in the gut microbiome of MASLD and identified microbial heterogeneity within intestinal compartments. Insights into sex-specific variations in gut microbiome composition could facilitate customised treatment strategies.
Keywords: Gut microbiome; MASLD; Metabolic syndrome; NAFLD; Sexual dimorphism.
Conflict of interest statement
The authors have no conflicts to disclose.
Figures






References
-
- Rinella ME, Lazarus JV, Ratziu V, Francque SM, Sanyal AJ, Kanwal F, et al. A multisociety Delphi consensus statement on new fatty liver disease nomenclature. J Hepatol. 2023;79:1542–1556. - PubMed
-
- Brunt EM, Wong VW, Nobili V, Day CP, Sookoian S, Maher JJ, et al. Nonalcoholic fatty liver disease. Nat Rev Dis Primers. 2015;1:15080. - PubMed
-
- Mouzaki M, Comelli EM, Arendt BM, Bonengel J, Fung SK, Fischer SE, et al. Intestinal microbiota in patients with nonalcoholic fatty liver disease. Hepatology. 2013;58:120–127. - PubMed
MeSH terms
Substances
Grants and funding
- Agencia Nacional de Promoción Científica y Tecnológica
- PICT 2018-889/Fondo para la Investigación Científica y Tecnológica
- PICT 2019-0528/Fondo para la Investigación Científica y Tecnológica
- PICT 2018-00620/Fondo para la Investigación Científica y Tecnológica
- PICT 2020-SerieA-0799/Fondo para la Investigación Científica y Tecnológica
LinkOut - more resources
Full Text Sources
Medical

