A Rare Noncoding Enhancer Variant in SCN5A Contributes to the High Prevalence of Brugada Syndrome in Thailand
- PMID: 39391988
- PMCID: PMC11670919
- DOI: 10.1161/CIRCULATIONAHA.124.069041
A Rare Noncoding Enhancer Variant in SCN5A Contributes to the High Prevalence of Brugada Syndrome in Thailand
Abstract
Background: Brugada syndrome (BrS) is a cardiac arrhythmia disorder that causes sudden death in young adults. Rare genetic variants in the SCN5A gene encoding the Nav1.5 sodium channel and common noncoding variants at this locus are robustly associated with the condition. BrS is particularly prevalent in Southeast Asia but the underlying ancestry-specific factors remain largely unknown.
Methods: Genome sequencing of BrS probands and population-matched controls from Thailand was performed to identify rare noncoding variants at the SCN5A-SCN10A locus that were enriched in patients with BrS. A likely causal variant was prioritized by computational methods and introduced into human induced pluripotent stem cell (hiPSC) lines using CRISPR-Cas9. The effect of the variant on SCN5A expression and Nav1.5 sodium channel current was then assessed in hiPSC-derived cardiomyocytes (hiPSC-CMs).
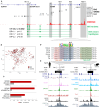
Results: A rare noncoding variant in an SCN5A intronic enhancer region was highly enriched in patients with BrS (detected in 3.9% of cases with a case-control odds ratio of 45.2). The variant affects a nucleotide conserved across all mammalian species and predicted to disrupt a Mef2 transcription factor binding site. Heterozygous introduction of the enhancer variant in hiPSC-CMs caused significantly reduced SCN5A expression from the variant-containing allele and a 30% reduction in Nav1.5-mediated sodium current density compared with isogenic controls, confirming its pathogenicity. Patients with the variant had severe phenotypes, with 89% experiencing cardiac arrest.
Conclusions: This is the first example of a functionally validated rare noncoding variant at the SCN5A locus and highlights how genome sequencing in understudied populations can identify novel disease mechanisms. The variant partly explains the increased prevalence of BrS in this region and enables the identification of at-risk variant carriers to reduce the burden of sudden cardiac death in Thailand.
Keywords: Asia, Southeastern; Brugada syndrome; genetics; noncoding genetic variant.
Conflict of interest statement
None.
Figures


References
-
- Behr ER, Ben-Haim Y, Ackerman MJ, Krahn AD, Wilde AAM. Brugada syndrome and reduced right ventricular outflow tract conduction reserve: a final common pathway? Eur Heart J. 2021;42:1073–1081. doi: 10.1093/eurheartj/ehaa1051 - PubMed
-
- Walsh R, Lahrouchi N, Tadros R, Kyndt F, Glinge C, Postema PG, Amin AS, Nannenberg EA, Ware JS, Whiffin N, et al. ; Nantes Referral Center for inherited cardiac arrhythmia. Enhancing rare variant interpretation in inherited arrhythmias through quantitative analysis of consortium disease cohorts and population controls. Genet Med. 2021;23:47–58. doi: 10.1038/s41436-020-00946-5 - PMC - PubMed
-
- Hosseini SM, Kim R, Udupa S, Costain G, Jobling R, Liston E, Jamal SM, Szybowska M, Morel CF, Bowdin S, et al. ; National Institutes of Health Clinical Genome Resource Consortium. Reappraisal of reported genes for sudden arrhythmic death. Circulation. 2018;138:1195–1205. doi: 10.1161/CIRCULATIONAHA.118.035070 - PMC - PubMed
-
- Wilde AAM, Semsarian C, Márquez MF, Shamloo AS, Ackerman MJ, Ashley EA, Sternick EB, Barajas-Martinez H, Behr ER, Bezzina CR, et al. ; Document Reviewers. European Heart Rhythm Association (EHRA)/Heart Rhythm Society (HRS)/Asia Pacific Heart Rhythm Society (APHRS)/Latin American Heart Rhythm Society (LAHRS) expert consensus statement on the state of genetic testing for cardiac diseases. Europace. 2022;24:1307–1367. doi: 10.1093/europace/euac030 - PMC - PubMed
-
- Barc J, Tadros R, Glinge C, Chiang DY, Jouni M, Simonet F, Jurgens SJ, Baudic M, Nicastro M, Potet F, et al. ; KORA-Study Group. Genome-wide association analyses identify new Brugada syndrome risk loci and highlight a new mechanism of sodium channel regulation in disease susceptibility. Nat Genet. 2022;54:232–239. doi: 10.1038/s41588-021-01007-6 - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

