Performance of a Protein Language Model for Variant Annotation in Cardiac Disease
- PMID: 39392163
- PMCID: PMC11935564
- DOI: 10.1161/JAHA.124.036921
Performance of a Protein Language Model for Variant Annotation in Cardiac Disease
Abstract
Background: Genetic testing is a cornerstone in the assessment of many cardiac diseases. However, variants are frequently classified as variants of unknown significance, limiting the utility of testing. Recently, the DeepMind group (Google) developed AlphaMissense, a unique artificial intelligence-based model, based on language model principles, for the prediction of missense variant pathogenicity. We aimed to report on the performance of AlphaMissense, accessed by VarCardio, an open web-based variant annotation engine, in a real-world cardiovascular genetics center.
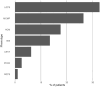
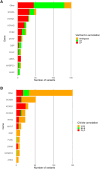
Methods and results: All genetic variants from an inherited arrhythmia program were examined using AlphaMissense via VarCard.io and compared with the ClinVar variant classification system, as well as another variant classification platform (Franklin by Genoox). The mutation reclassification rate and genotype-phenotype concordance were examined for all variants in the study. We included 266 patients with heritable cardiac diseases, harboring 339 missense variants. Of those, 230 (67.8%) were classified by ClinVar as either variants of unknown significance or nonclassified. Using VarCard.io, 198 variants of unknown significance (86.1%, 95% CI, 80.9-90.3) were reclassified to either likely pathogenic or likely benign. The reclassification rate was significantly higher for VarCard.io than for Franklin (86.1% versus 34.8%, P<0.001). Genotype-phenotype concordance was highly aligned using VarCard.io predictions, at 95.9% (95% CI, 92.8-97.9) concordance rate. For 109 variants classified as pathogenic, likely pathogenic, benign, or likely benign by ClinVar, concordance with VarCard.io was high (90.5%).
Conclusions: AlphaMissense, accessed via VarCard.io, may be a highly efficient tool for cardiac genetic variant interpretation. The engine's notable performance in assessing variants that are classified as variants of unknown significance in ClinVar demonstrates its potential to enhance cardiac genetic testing.
Keywords: AlphaMissense; VarCard.Io; artificial intelligence; genetic testing; genotype; phenotype; variants of unknown significance.
Figures

References
-
- Mazzaccara C, Lombardi R, Mirra B, Barretta F, Esposito MV, Uomo F, Caiazza M, Monda E, Losi MA, Limongelli G, et al. Next‐generation sequencing gene panels in inheritable cardiomyopathies and channelopathies: prevalence of pathogenic variants and variants of unknown significance in uncommon genes. Biomolecules. 2022;12:1417. doi: 10.3390/biom12101417 - DOI - PMC - PubMed
-
- van Lint FHM, Mook ORF, Alders M, Bikker H, Lekanne D, Deprez RH, Christiaans I. Large next‐generation sequencing gene panels in genetic heart disease: yield of pathogenic variants and variants of unknown significance. Neth Heart J. 2019;27:304–309. doi: 10.1007/s12471-019-1250-5 - DOI - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources

