Implementation and validation of face de-identification (de-facing) in ADNI4
- PMID: 39392215
- PMCID: PMC11567833
- DOI: 10.1002/alz.14303
Implementation and validation of face de-identification (de-facing) in ADNI4
Abstract
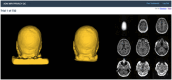
Introduction: Recent technological advances have increased the risk that de-identified brain images could be re-identified from face imagery. The Alzheimer's Disease Neuroimaging Initiative (ADNI) is a leading source of publicly available de-identified brain imaging, who quickly acted to protect participants' privacy.
Methods: An independent expert committee evaluated 11 face-deidentification ("de-facing") methods and selected four for formal testing.
Results: Effects of de-facing on brain measurements were comparable across methods and sufficiently small to recommend de-facing in ADNI. The committee ultimately recommended mri_reface for advantages in reliability, and for some practical considerations. ADNI leadership approved the committee's recommendation, beginning in ADNI4.
Discussion: ADNI4 de-faces all applicable brain images before subsequent pre-processing, analyses, and public release. Trained analysts inspect de-faced images to confirm complete face removal and complete non-modification of brain. This paper details the history of the algorithm selection process and extensive validation, then describes the production workflows for de-facing in ADNI.
Highlights: ADNI is implementing "de-facing" of MRI and PET beginning in ADNI4. "De-facing" alters face imagery in brain images to help protect privacy. Four algorithms were extensively compared for ADNI and mri_reface was chosen. Validation confirms mri_reface is robust and effective for ADNI sequences. Validation confirms mri_reface negligibly affects ADNI brain measurements.
Keywords: ADNI; anonymization; de‐facing; de‐identification; face recognition.
© 2024 The Author(s). Alzheimer's & Dementia published by Wiley Periodicals LLC on behalf of Alzheimer's Association.
Conflict of interest statement
Christopher G. Schwarz: receives research funding from the NIH, related and unrelated to this work. Mark Choe: reports no disclosures. Stephanie Rossi: reports no disclosures. Sandhitsu R. Das: reports no disclosures. Ranjit Ittyerah: reports no disclosures. Evan Fletcher: reports no disclosures. Pauline Maillard: reports no disclosures. Baljeet Singh: reports no disclosures. Danielle J. Harvey: receives research funding from the NIH, related and unrelated to this work, serves as statistical advisor to PLOS ONE. Ian B. Malone: is supported by grants to his institution from NIH and is an employee of the Dementia Research Centre which is supported by Alzheimer's Research UK, Brain Research Trust, and The Wolfson Foundation. Lloyd Prosser: works for the Dementia Research Centre (DRC) which is supported by: Alzheimer's Research UK; Brain Research Trust; the Wolfson Foundation. L. Prosser reports no additional disclosures to those that support the DRC. Matthew L. Senjem: reports no disclosures. Leonard C. Matoush: reports no disclosures. Chadwick P. Ward: reports no disclosures. Carl M. Prakaashana: reports no disclosures. Susan M. Landau: receives research funding from the NIH, related and unrelated to this work, is on the DSMB/SAB for KeifeRx and the NIH IPAT study, has received speaking honoraria from Eisai and IMPACT‐AD, has consulted for Banner Health, Vaccinex, and has received travel funding and other research support from IMPACT AD and the Alzheimer's Association. Robert A. Koeppe: reports no disclosures. JiaQie Lee: reports no disclosures. Charles DeCarli: reports no disclosures. Michael W. Weiner: serves on Editorial Boards for Alzheimer's & Dementia, and the Journal for Prevention of Alzheimer's Disease (JPAD). He has served on Advisory Boards for Acumen Pharmaceutical, Alzheon, Inc., Amsterdam UMC; MIRIADE, Cerecin, Merck Sharp & Dohme Corp., NC Registry for Brain Health, and REGEnLIFE. He also serves on the USC ACTC grant which receives funding from Eisai. He has provided consulting to Boxer Capital, LLC, Cerecin, Inc., Clario, Dementia Society of Japan, Dolby Family Ventures, Eisai, Guidepoint, Health and Wellness Partners, Indiana University, LCN Consulting, MEDA Corp., Merck Sharp & Dohme Corp., NC Registry for Brain Health, Prova Education, T3D Therapeutics, University of Southern California (USC), and WebMD. He has acted as a speaker/lecturer for China Association for Alzheimer's Disease (CAAD) and Taipei Medical University, as well as a speaker/lecturer with academic travel funding provided by: AD/PD Congress, Amsterdam UMC, Cleveland Clinic, CTAD Congress, Foundation of Learning; Health Society (Japan), Kenes, U. Penn, U. Toulouse, Japan Society for Dementia Research, Korean Dementia Society, Merck Sharp & Dohme Corp., National Center for Geriatrics and Gerontology (NCGG; Japan), University of Southern California (USC). He holds stock options with Alzeca, Alzheon, Inc., ALZPath, Inc., and Anven. Dr Weiner received support for his research from the following funding sources: National Institutes of Health (NIH)/NINDS/National Institute on Aging (NIA), Department of Defense (DOD), California Department of Public Health (CDPH), University of Michigan, Siemens, Biogen, Hillblom Foundation, Alzheimer's Association, Johnson & Johnson, Kevin and Connie Shanahan, GE, VUmc, Australian Catholic University (HBI‐BHR), The Stroke Foundation, and the Veterans Administration. Clifford R. Jack, Jr.: reports no disclosures. William J. Jagust: Jagust receives research funding from the NIH and Genentech, has consulted for Lilly, Biogen, Clario and Eisai and holds equity in Molecular Medicine and Optoceutics. Paul A. Yushkevich: receives research funding from the NIH, related and unrelated to this work. Duygu Tosun: receives research funding from the National Institute on Aging (NIH). Author disclosures are available in the supporting information.
Figures






References
-
- Provenzano D, Loew MH, Goyal S, Rao YJ. Unmasking a privacy concern: potential identification of patients in an immobilization mask from 3‐dimensional reconstructions of simulation computed tomography. Practical Radiation Oncology. 2021:S1879850021002356. doi:10.1016/j.prro.2021.09.006 - DOI - PubMed

