Functional RNAi Screening Identifies G2/M and Kinetochore Components as Modulators of TNFα/NF-κB Prosurvival Signaling in Head and Neck Squamous Cell Carcinoma
- PMID: 39392349
- PMCID: PMC11541648
- DOI: 10.1158/2767-9764.CRC-24-0274
Functional RNAi Screening Identifies G2/M and Kinetochore Components as Modulators of TNFα/NF-κB Prosurvival Signaling in Head and Neck Squamous Cell Carcinoma
Abstract
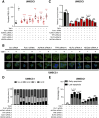
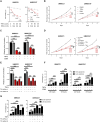
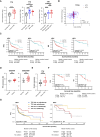
Here, RNAi library screening reveals that multiple G2/M and kinetochore components, including TTK/monopolar spindle 1, modulate TNFα-induced NF-κB activation, cell survival, and genotoxicity, underscoring their potential importance as therapeutic targets in HNSCC.
©2024 The Authors; Published by the American Association for Cancer Research.
Conflict of interest statement
A.D. Saleh reports other from miRecule, Inc. outside the submitted work. S.E. Martin reports other from Genentech outside the submitted work. C. Van Waes reports grants from NIH during the conduct of the study, as well as personal fees from miRecule, Inc. outside the submitted work. No disclosures were reported by the other authors.
Figures




References
-
- Kim HJ, Hawke N, Baldwin AS. NF-kappaB and IKK as therapeutic targets in cancer. Cell Death Differ 2006;13:738–47. - PubMed
-
- Allen CT, Ricker JL, Chen Z, Van Waes C. Role of activated nuclear factor-kappaB in the pathogenesis and therapy of squamous cell carcinoma of the head and neck. Head Neck 2007;29:959–71. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

