Deubiquitylase USP52 Promotes Bladder Cancer Progression by Modulating Ferroptosis through Stabilizing SLC7A11/xCT
- PMID: 39392373
- PMCID: PMC11615784
- DOI: 10.1002/advs.202403995
Deubiquitylase USP52 Promotes Bladder Cancer Progression by Modulating Ferroptosis through Stabilizing SLC7A11/xCT
Abstract
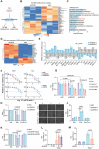
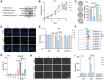
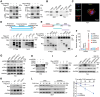
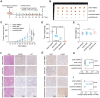
Bladder cancer (BLCA) is a prevalent cancer with high case-fatality rates and a substantial economic burden worldwide. Understanding its molecular underpinnings to guide clinical management is crucial. Ferroptosis, a recently described non-apoptotic form of cell death, is initiated by the lethal accumulation of iron-dependent lipid peroxidation products. Despite growing interest, the roles and vulnerabilities determining ferroptosis sensitivity in BLCA remain unclear. Re-analysis of single-cell RNA data reveals a decrease in high-ferroptosis cancer cells as BLCA advances. USP52/PAN2 is identified as a key regulator of ferroptosis in BLCA through an unbiased siRNA screen targeting 96 deubiquitylases (DUBs). Functionally, USP52 depletion impedes glutathione (GSH) synthesis by promoting xCT protein degradation, increasing lipid peroxidation and ferroptosis susceptibility, thus suppressing BLCA progression. Mechanistically, USP52 interacts with xCT and enzymatically cleaves the K48-conjugated ubiquitin chains at K4 and K12, enhancing its protein stability. Clinical BLCA samples demonstrate a positive correlation between USP52 and xCT expression, with high USP52 levels associated with aggressive disease progression and poor prognosis. In vivo, USP52 depletion combined with ferroptosis triggers imidazole ketone Erastin (IKE) synergistically restrains BLCA progression by inducing ferroptosis. These findings elucidate the role of the USP52-xCT axis in BLCA and highlight the therapeutic potential of targeting USP52 and ferroptosis inducers in BLCA.
Keywords: SLC7A11/xCT; USP52; bladder cancer; deubiquitylases; ferroptosis.
© 2024 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Sung H., Ferlay J., Siegel R. L., Laversanne M., Soerjomataram I., Jemal A., Bray F., CA Cancer J. Clin. 2021, 71, 209. - PubMed
-
- van Hoogstraten L. M. C., Vrieling A., van der Heijden A. G., Kogevinas M., Richters A., Kiemeney L. A., Nat. Rev. Clin. Oncol. 2023, 20, 287. - PubMed
-
- a) Dixon S. J., Lemberg K. M., Lamprecht M. R., Skouta R., Zaitsev E. M., Gleason C. E., Patel D. N., Bauer A. J., Cantley A. M., Yang W. S., Morrison B. 3rd, Stockwell B. R., Cell 2012, 149, 1060; - PMC - PubMed
- b) Xie Y., Hou W., Song X., Yu Y., Huang J., Sun X., Kang R., Tang D., Cell Death Differ. 2016, 23, 369; - PMC - PubMed
- c) Stockwell B. R., Friedmann Angeli J. P., Bayir H., Bush A. I., Conrad M., Dixon S. J., Fulda S., Gascón S., Hatzios S. K., Kagan V. E., Noel K., Jiang X., Linkermann A., Murphy M. E., Overholtzer M., Oyagi A., Pagnussat G. C., Park J., Ran Q., Rosenfeld C. S., Salnikow K., Tang D., Torti F. M., Torti S. V., Toyokuni S., Woerpel K. A., Zhang D. D., Cell 2017, 171, 273. - PMC - PubMed
MeSH terms
Substances
Grants and funding
- 82472733/National Natural Science Foundation of China
- 2042022dx0003/Fundamental Research Funds for the Central Universities
- PTPP2024001/Research Fund of Zhongnan Hospital of Wuhan University
- SWYBK01-03/Research Fund of Zhongnan Hospital of Wuhan University
- RLYC2024001001/Research Fund of Zhongnan Hospital of Wuhan University
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
