Protocol for CRISPR-Cas9 genome editing of a swine cell line via electroporation
- PMID: 39392744
- PMCID: PMC11735999
- DOI: 10.1016/j.xpro.2024.103385
Protocol for CRISPR-Cas9 genome editing of a swine cell line via electroporation
Abstract
Genome editing technology is being used in animals for a variety of purposes, including improvement of animal and public health outcomes. Characterization of genome editing reagents and anticipated genomic alterations is an essential step toward the development of an edited animal. Here, we present a protocol for genome editing in the swine testicular (ST) cell line. We describe steps for evaluating CRISPR-Cas9 complex functionality in vitro, delivering editing molecules into cells by transfection, and assessing target editing via Sanger sequencing.
Keywords: Biotechnology and bioengineering; CRISPR; Cell-based Assays.
Published by Elsevier Inc.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures
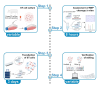


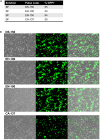


References
-
- Burger B.T., Beaton B.P., Campbell M.A., Brett B.T., Rohrer M.S., Plummer S., Barnes D., Jiang K., Naswa S., Lange J., et al. Generation of a Commercial-Scale Founder Population of Porcine Reproductive and Respiratory Syndrome Virus Resistant Pigs Using CRISPR-Cas. CRISPR J. 2024;7:12–28. doi: 10.1089/crispr.2023.0061. - DOI - PubMed
-
- Moretti A., Fonteyne L., Giesert F., Hoppmann P., Meier A.B., Bozoglu T., Baehr A., Schneider C.M., Sinnecker D., Klett K., et al. Somatic gene editing ameliorates skeletal and cardiac muscle failure in pig and human models of Duchenne muscular dystrophy. Nat. Med. 2020;26:207–214. doi: 10.1038/s41591-019-0738-2. - DOI - PMC - PubMed
-
- Tu C.F., Chuang C.K., Hsiao K.H., Chen C.H., Chen C.M., Peng S.H., Su Y.H., Chiou M.T., Yen C.H., Hung S.W., et al. Lessening of porcine epidemic diarrhoea virus susceptibility in piglets after editing of the CMP-N-glycolylneuraminic acid hydroxylase gene with CRISPR/Cas9 to nullify N-glycolylneuraminic acid expression. PLoS One. 2019;14 doi: 10.1371/journal.pone.0217236. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

