Interactions of Polychlorinated Biphenyls and Their Metabolites with the Brain and Liver Transcriptome of Female Mice
- PMID: 39392776
- PMCID: PMC11587508
- DOI: 10.1021/acschemneuro.4c00367
Interactions of Polychlorinated Biphenyls and Their Metabolites with the Brain and Liver Transcriptome of Female Mice
Abstract
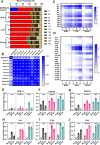
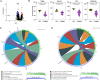
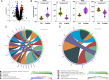
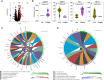
Exposure to polychlorinated biphenyls (PCBs) is linked to neurotoxic effects. This study aims to close knowledge gaps regarding the specific modes of action of PCBs in female C57BL/6J mice (>6 weeks) orally exposed for 7 weeks to a human-relevant PCB mixture (MARBLES mix) at 0, 0.1, 1, and 6 mg/kg body weight/day. PCB and hydroxylated PCB (OH-PCBs) levels were quantified in the brain, liver, and serum; RNA sequencing was performed in the striatum, prefrontal cortex, and liver, and metabolomic analyses were performed in the striatum. Profiles of PCBs but not their hydroxylated metabolites were similar in all tissues. In the prefrontal cortex, PCB exposure activated the oxidative phosphorylation respiration pathways, while suppressing the axon guidance pathway. PCB exposure significantly changed the expression of genes associated with neurodevelopmental and neurodegenerative diseases in the striatum, impacting pathways like growth hormone synthesis and dendrite development. PCBs did not affect the striatal metabolome. In contrast to the liver, which showed activation of metabolic processes following PCB exposure and the induction of cytochrome P450 enzymes, the expression of xenobiotic processing genes was not altered by PCB exposure in either brain region. Network analysis revealed complex interactions between individual PCBs (e.g., PCB28 [2,4,4'-trichlorobiphenyl]) and their hydroxylated metabolites and specific differentially expressed genes (DEGs), underscoring the need to characterize the association between specific PCBs and DEGs. These findings enhance the understanding of PCB neurotoxic mechanisms and their potential implications for human health.
Keywords: RNA sequencing; metabolomics; multiomics; network analysis; neurotoxicity; polychlorinated biphenyls.
Conflict of interest statement
The authors declare no competing financial interest.
Figures



References
-
- ATSDR . Toxicological profile for polychlorinated biphenyls. 2000. - PubMed
-
- EPA Polychlorinated Biphenyls (PCBs) . Inadvertent PCBs. https://www.epa.gov/pcbs/inadvertent-pcbs.
-
- Carlson L. M.; Christensen K.; Sagiv S. K.; Rajan P.; Klocke C. R.; Lein P. J.; Coffman E.; Shaffer R. M.; Yost E. E.; Arzuaga X.; Factor-Litvak P.; Sergeev A.; Toborek M.; Bloom M. S.; Trgovcich J.; Jusko T. A.; Robertson L.; Meeker J.; Keating A. F.; Blain R.; Silva R.; Snow S.; Lin C.; Shipkowski K.; Ingle B.; Lehmann G. M. A systematic evidence map for the evaluation of noncancer health effects and exposures to polychlorinated biphenyl mixtures. Environ. Res. 2023, 220, 115148. 10.1016/j.envres.2022.115148. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

