Transcriptomic analyses of host-virus interactions during in vitro infection with wild-type and glycoprotein g-deficient (ΔgG) strains of ILTV in primary and continuous cell cultures
- PMID: 39392810
- PMCID: PMC11469545
- DOI: 10.1371/journal.pone.0311874
Transcriptomic analyses of host-virus interactions during in vitro infection with wild-type and glycoprotein g-deficient (ΔgG) strains of ILTV in primary and continuous cell cultures
Abstract
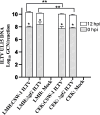
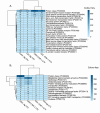
Infectious laryngotracheitis (ILT) remains a significant concern for the poultry industry worldwide due to its impact on animal welfare and its substantial economic consequences. The disease is caused by the alphaherpesvirus, infectious laryngotracheitis virus (ILTV). This study investigated in vitro host-virus interactions of a glycoprotein G (gG) deletion mutant vaccine strain of ILTV (ΔgG ILTV), and its parent wild-type strain (CSW-1 ILTV). Inoculations were performed separately for the two strains of ILTV using both a primary (chicken embryonic kidney, CEK) and a continuous culture (leghorn male hepatoma, LMH) of chicken cells. Transcriptome analysis was performed at 12 hours post infection. Each cell-type displayed distinct effects on host and viral gene transcription, with a greater number of viral and host genes differentially transcribed in CEK cells and LMH cells, respectively. Both cell-types infected with either strain demonstrated enrichment of pathways related to signalling, and gene ontologies (GO) associated with chemotaxis. Infection with either strain upregulated both SOCS proteins and certain proto-oncogenes, which may contribute to prolonged viral persistence by promoting immunosuppression and preventing apoptosis, respectively. Patterns of gene transcription related to cytokines, chemokines, endosomal TLRs, and interferon responses, as well as pathways associated with histone acetylation, transport, and extracellular matrix organization were similar within each cell type, regardless of the viral strain. In CEK cells, GO terms and pathways were downregulated uniquely after CSW-1 ILTV infection, indicating a viral-strain specific effect in this cell-type. Overall, this study highlights that the observed differences in host and ILTV gene transcription in vitro were more strongly influenced by the cell-types used rather than the presence or absence of gG. This underscores the importance of cell-line selection in studying host-virus interactions and interpreting experimental results.
Copyright: © 2024 Gopakumar et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Bagust TJ, Jones RC, Guy JS. Avian infectious laryngotracheitis. Revue scientifique et technique. 2000;19:483–92. - PubMed
-
- Prideaux CTK K. Johnson, M.A. Sheppard, M. and Fahey, K.J. Infectious laryngotracheitis virus growth, DNA replication, and protein synthesis. Archives of Virology. 1992(123):181–92. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

