Environmental predictors impact microbial-based postmortem interval (PMI) estimation models within human decomposition soils
- PMID: 39392823
- PMCID: PMC11469530
- DOI: 10.1371/journal.pone.0311906
Environmental predictors impact microbial-based postmortem interval (PMI) estimation models within human decomposition soils
Abstract
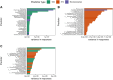
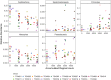
Microbial succession has been suggested to supplement established postmortem interval (PMI) estimation methods for human remains. Due to limitations of entomological and morphological PMI methods, microbes are an intriguing target for forensic applications as they are present at all stages of decomposition. Previous machine learning models from soil necrobiome data have produced PMI error rates from two and a half to six days; however, these models are built solely on amplicon sequencing of biomarkers (e.g., 16S, 18S rRNA genes) and do not consider environmental factors that influence the presence and abundance of microbial decomposers. This study builds upon current research by evaluating the inclusion of environmental data on microbial-based PMI estimates from decomposition soil samples. Random forest regression models were built to predict PMI using relative taxon abundances obtained from different biological markers (bacterial 16S, fungal ITS, 16S-ITS combined) and taxonomic levels (phylum, class, order, OTU), both with and without environmental predictors (ambient temperature, soil pH, soil conductivity, and enzyme activities) from 19 deceased human individuals that decomposed on the soil surface (Tennessee, USA). Model performance was evaluated by calculating the mean absolute error (MAE). MAE ranged from 804 to 997 accumulated degree hours (ADH) across all models. 16S models outperformed ITS models (p = 0.006), while combining 16S and ITS did not improve upon 16S models alone (p = 0.47). Inclusion of environmental data in PMI prediction models had varied effects on MAE depending on the biological marker and taxonomic level conserved. Specifically, inclusion of the measured environmental features reduced MAE for all ITS models, but improved 16S models at higher taxonomic levels (phylum and class). Overall, we demonstrated some level of predictability in soil microbial succession during human decomposition, however error rates were high when considering a moderate population of donors.
Copyright: © 2024 Mason et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





Similar articles
-
Unveiling the Forensic Potential of Oral and Nasal Microbiota in Post-Mortem Interval Estimation.Int J Mol Sci. 2025 Apr 6;26(7):3432. doi: 10.3390/ijms26073432. Int J Mol Sci. 2025. PMID: 40244278 Free PMC article.
-
Microbial succession patterns for postmortem interval estimation in decomposed mouse cadavers: A comparative study of mechanical asphyxia and hemorrhagic shock.J Forensic Sci. 2025 Sep;70(5):1892-1907. doi: 10.1111/1556-4029.70108. Epub 2025 Jun 16. J Forensic Sci. 2025. PMID: 40524283
-
Metabarcoding to investigate changes in soil microbial communities within forensic burial contexts.Forensic Sci Int Genet. 2019 Mar;39:73-85. doi: 10.1016/j.fsigen.2018.12.002. Epub 2018 Dec 12. Forensic Sci Int Genet. 2019. PMID: 30594064
-
From microbial data to forensic insights: systematic review of machine learning models for PMI estimation.Forensic Sci Med Pathol. 2025 Apr 21. doi: 10.1007/s12024-025-01002-x. Online ahead of print. Forensic Sci Med Pathol. 2025. PMID: 40259168 Review.
-
Application of High-throughput Sequencing in Researches of Cadaveric Microorganisms and Postmortem Interval Estimation.Fa Yi Xue Za Zhi. 2018 Oct;34(5):475-481. doi: 10.12116/j.issn.1004-5619.2018.05.004. Epub 2018 Oct 25. Fa Yi Xue Za Zhi. 2018. PMID: 30468048 Review. Chinese, English.
Cited by
-
Development of Mathematical Models Using circRNA Combinations (circTulp4, circSlc8a1, and circStrn3) in Mouse Brain Tissue for Postmortem Interval Estimation.Int J Mol Sci. 2025 May 8;26(10):4495. doi: 10.3390/ijms26104495. Int J Mol Sci. 2025. PMID: 40429639 Free PMC article.
-
Advances in Forensic Entomotoxicology for Decomposed Corpses: A Review.Insects. 2025 Jul 21;16(7):744. doi: 10.3390/insects16070744. Insects. 2025. PMID: 40725374 Free PMC article. Review.
-
Circular RNA circFat3 as a biomarker for construction of postmortem interval Estimation models in mouse brain tissues at multiple temperatures.Sci Rep. 2025 Jul 1;15(1):21577. doi: 10.1038/s41598-025-07998-0. Sci Rep. 2025. PMID: 40593252 Free PMC article.
References
-
- Howard GT, Duos B, Watson-Horzelski EJ. Characterization of the soil microbial community associated with the decomposition of a swine carcass. International Biodeterioration & Biodegradation. 2010;64(4):300–304. doi: 10.1016/j.ibiod.2010.02.006 - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

