Comprehensive mutational profiling identifies new driver events in cutaneous leiomyosarcoma
- PMID: 39392932
- PMCID: PMC11758588
- DOI: 10.1093/bjd/ljae386
Comprehensive mutational profiling identifies new driver events in cutaneous leiomyosarcoma
Abstract
Background: Cutaneous leiomyosarcoma (cLMS) is a rare soft-tissue neoplasm, showing smooth muscle differentiation, that arises from the mesenchymal cells of the dermis. To date, genetic investigation of these tumours has involved studies with small sample sizes and limited analyses that identified recurrent somatic mutations in RB1 and TP53, copy number gain of MYOCD and IGF1R, and copy number loss of PTEN.
Objectives: To better understand the molecular pathogenesis of cLMS, we comprehensively explored the mutational landscape of these rare tumours to identify candidate driver events.
Methods: In this retrospective, multi-institutional study, we performed whole-exome sequencing and RNA sequencing in 38 cases of cLMS.
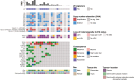
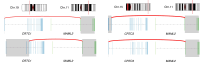
Results: TP53 and RB1 were identified as significantly mutated and thus represent validated driver genes of cLMS. COSMIC mutational signatures SBS7a/b and DBS1 were recurrent; thus, ultraviolet light exposure may be an aetiological factor driving cLMS. Analysis of significantly recurrent somatic copy number alterations, which represent candidate driver events, found focal (< 10 Mb) deletions encompassing TP53 and KDM6B, and amplifications encompassing ZMYM2, MYOCD, MAP2K4 and NCOR1. A larger (24 Mb) recurrent deletion encompassing CYLD was also identified as significant. Significantly recurrent broad copy number alterations, involving at least half of a chromosome arm, included deletions of 6p/q, 10p/q, 11q, 12q, 13q and 16p/q, and amplification of 15q. Notably PTEN is located on 10q, RB1 on 13q and IGFR1 on 15q. Fusion gene analysis identified recurrent CRTC1/CRTC3::MAML2 fusions, as well as many novel fusions in individual samples.
Conclusions: Our analysis of the largest number of cases of cLMS to date highlights the importance of large cohort sizes and exploration beyond small targeted gene panels when performing molecular analyses, as it allowed a comprehensive exploration of the mutational landscape of these tumours and identification of novel candidate driver events. It also uniquely afforded the opportunity to compare the molecular phenotype of cLMS with LMS of other tissue types, such as uterine and soft-tissue LMS. Given that molecular profiling has resulted in the development of novel targeted treatment approaches for uterine and soft-tissue LMS, our study now allows the same opportunities to become available for patients with cLMS.
Plain language summary
Cutaneous leiomyosarcoma (or ‘cLMS’ for short) is a type of skin tumour. It commonly appears as a painful nodule on the trunk, legs or arms. Surgery to remove it is recommended, but it can grow back and may spread to other organs. For this reason, we need to understand more about cLMS to try to find other treatments. Analysing the genetic material in tumour cells lets us know which genes have been mutated or altered. This is important as these mutations or alterations can play a role in the development of a tumour or its progression. Recording all the mutations and alterations found in a tumour is crucial to understanding the biology of the disease. It is also important in finding ways to diagnose the disease, predicting how it will progress and finding treatments for it. Up to now, only two studies have analysed the genetic material in cLMS tumours. Samples were only available from small groups of people and fewer than 100 genes were investigated. To study the genetic changes in cLMS and identify the key events that cause them, we analysed all the genes in the DNA of a large number of people with cLMS. We confirmed that two genes (called ‘TP53’ and ‘RB1’) could be key to the development of cLMS. We also found new possible causes of cLMS, including exposure to sunlight, changes in chromosomes and some gene fusions. These findings give us a better understanding of the biology of cLMS.
© The Author(s) 2024. Published by Oxford University Press on behalf of British Association of Dermatologists. All rights reserved. For commercial re-use, please contact reprints@oup.com for reprints and translation rights for reprints. All other permissions can be obtained through our RightsLink service via the Permissions link on the article page on our site—for further information please contact journals.permissions@oup.com.
Conflict of interest statement
Conflicts of interest: The authors declare no conflicts of interest.
Figures
References
-
- Helbig D, Dippel E, Erdmann M et al. S1-guideline cutaneous and subcutaneous leiomyosarcoma. J Dtsch Dermatol Ges 2023; 21:555–63. - PubMed
-
- Bresler SC, Gosnell HL, Ko JS et al. Subcutaneous leiomyosarcoma: an aggressive malignancy portending a significant risk of metastasis and death. Am J Surg Pathol 2023; 47:1417–24. - PubMed
-
- Fields JP, Helwig EB. Leiomyosarcoma of the skin and subcutaneous tissue. Cancer 1981; 47:156–69. - PubMed
-
- Kraft S, Fletcher CD. Atypical intradermal smooth muscle neoplasms: clinicopathologic analysis of 84 cases and a reappraisal of cutaneous “leiomyosarcoma”. Am J Surg Pathol 2011; 35:599–607. - PubMed
-
- Massi D, Franchi A, Alos L et al. Primary cutaneous leiomyosarcoma: clinicopathological analysis of 36 cases. Histopathology 2010; 56:251–62. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

