N-cadherin crosstalk with integrin weakens the molecular clutch in response to surface viscosity
- PMID: 39394209
- PMCID: PMC11479646
- DOI: 10.1038/s41467-024-53107-6
N-cadherin crosstalk with integrin weakens the molecular clutch in response to surface viscosity
Abstract
Mesenchymal stem cells (MSCs) interact with their surroundings via integrins, which link to the actin cytoskeleton and translate physical cues into biochemical signals through mechanotransduction. N-cadherins enable cell-cell communication and are also linked to the cytoskeleton. This crosstalk between integrins and cadherins modulates MSC mechanotransduction and fate. Here we show the role of this crosstalk in the mechanosensing of viscosity using supported lipid bilayers as substrates of varying viscosity. We functionalize these lipid bilayers with adhesion peptides for integrins (RGD) and N-cadherins (HAVDI), to demonstrate that integrins and cadherins compete for the actin cytoskeleton, leading to an altered MSC mechanosensing response. This response is characterised by a weaker integrin adhesion to the environment when cadherin ligation occurs. We model this competition via a modified molecular clutch model, which drives the integrin/cadherin crosstalk in response to surface viscosity, ultimately controlling MSC lineage commitment.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
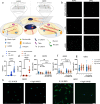
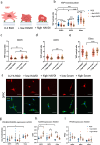
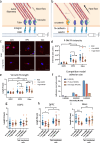
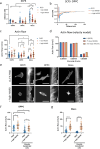
References
Publication types
MeSH terms
Substances
Grants and funding
- 101054728/EC | EU Framework Programme for Research and Innovation H2020 | H2020 Priority Excellent Science | H2020 European Research Council (H2020 Excellent Science - European Research Council)
- EP/X033554/1/RCUK | Engineering and Physical Sciences Research Council (EPSRC)
- MR/S005412/1/MRC_/Medical Research Council/United Kingdom
- 874889/EC | Horizon 2020 Framework Programme (EU Framework Programme for Research and Innovation H2020)
- RGS/R1/231400/Royal Society
LinkOut - more resources
Full Text Sources
Research Materials

