miR-210 is essential to retinal homeostasis in fruit flies and mice
- PMID: 39394614
- PMCID: PMC11468086
- DOI: 10.1186/s13062-024-00542-6
miR-210 is essential to retinal homeostasis in fruit flies and mice
Abstract
Background: miR-210 is one of the most evolutionarily conserved microRNAs. It is known to be involved in several physiological and pathological processes, including response to hypoxia, angiogenesis, cardiovascular diseases and cancer. Recently, new roles of this microRNA are emerging in the context of eye and visual system homeostasis. Recent studies in Drosophila melanogaster unveiled that the absence of miR-210 leads to a progressive retinal degeneration characterized by the accumulation of lipid droplets and disruptions in lipid metabolism. However, the possible conservation of miR-210 knock-out effect in the mammalian retina has yet to be explored.
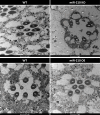
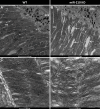
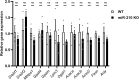
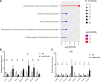
Results: We further investigated lipid anabolism and catabolism in miR-210 knock-out (KO) flies, uncovering significant alterations in gene expression within these pathways. Additionally, we characterized the retinal morphology of flies overexpressing (OE) miR-210, which was not affected by the increased levels of the microRNA. For the first time, we also characterized the retinal morphology of miR-210 KO and OE mice. Similar to flies, miR-210 OE did not affect retinal homeostasis, whereas miR-210 KO mice exhibited photoreceptor degeneration. To explore other potential parallels between miR-210 KO models in flies and mice, we examined lipid metabolism, circadian behaviour, and retinal transcriptome in mice, but found no similarities. Specifically, RNA-seq confirmed the lack of involvement of lipid metabolism in the mice's pathological phenotype, revealing that the differentially expressed genes were predominantly associated with chloride channel activity and extracellular matrix homeostasis. Simultaneously, transcriptome analysis of miR-210 KO fly brains indicated that the observed alterations extend beyond the eye and may be linked to neuronal deficiencies in signal detection and transduction.
Conclusions: We provide the first morphological characterization of the retina of miR-210 KO and OE mice, investigating the role of this microRNA in mammalian retinal physiology and exploring potential parallels with phenotypes observed in fly models. Although the lack of similarities in lipid metabolism, circadian behaviour, and retinal transcriptome in mice suggests divergent mechanisms of retinal degeneration between the two species, transcriptome analysis of miR-210 KO fly brains indicates the potential existence of a shared upstream mechanism contributing to retinal degeneration in both flies and mammals.
Keywords: Drosophila melanogaster; Mus musculus; Chloride channels; Circadian behaviour; Extracellular matrix; Lipid metabolism; Photoreceptor degeneration; Retina; Signal transduction; miR-210.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

