A corpus of GA4GH phenopackets: Case-level phenotyping for genomic diagnostics and discovery
- PMID: 39394689
- PMCID: PMC11564936
- DOI: 10.1016/j.xhgg.2024.100371
A corpus of GA4GH phenopackets: Case-level phenotyping for genomic diagnostics and discovery
Abstract
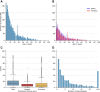
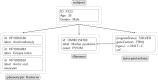
The Global Alliance for Genomics and Health (GA4GH) Phenopacket Schema was released in 2022 and approved by ISO as a standard for sharing clinical and genomic information about an individual, including phenotypic descriptions, numerical measurements, genetic information, diagnoses, and treatments. A phenopacket can be used as an input file for software that supports phenotype-driven genomic diagnostics and for algorithms that facilitate patient classification and stratification for identifying new diseases and treatments. There has been a great need for a collection of phenopackets to test software pipelines and algorithms. Here, we present Phenopacket Store. Phenopacket Store v.0.1.19 includes 6,668 phenopackets representing 475 Mendelian and chromosomal diseases associated with 423 genes and 3,834 unique pathogenic alleles curated from 959 different publications. This represents the first large-scale collection of case-level, standardized phenotypic information derived from case reports in the literature with detailed descriptions of the clinical data and will be useful for many purposes, including the development and testing of software for prioritizing genes and diseases in diagnostic genomics, machine learning analysis of clinical phenotype data, patient stratification, and genotype-phenotype correlations. This corpus also provides best-practice examples for curating literature-derived data using the GA4GH Phenopacket Schema.
Keywords: global alliance for genomics and health; human phenotype ontology; phenopacket schema.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests M.A.H. is a founder of Alamya Health. M.J.B. and J.X.C. are the Editor-in-Chief and Deputy Editor of HGG Advances, respectively, and were recused from the editorial handling of this manuscript.
Figures
Update of
-
A corpus of GA4GH Phenopackets: case-level phenotyping for genomic diagnostics and discovery.medRxiv [Preprint]. 2024 May 29:2024.05.29.24308104. doi: 10.1101/2024.05.29.24308104. medRxiv. 2024. Update in: HGG Adv. 2025 Jan 09;6(1):100371. doi: 10.1016/j.xhgg.2024.100371. PMID: 38854034 Free PMC article. Updated. Preprint.
References
-
- Putman T.E., Schaper K., Matentzoglu N., Rubinetti V.P., Alquaddoomi F.S., Cox C., Caufield J.H., Elsarboukh G., Gehrke S., Hegde H., et al. The Monarch Initiative in 2024: an analytic platform integrating phenotypes, genes and diseases across species. Nucleic Acids Res. 2024;52:D938–D949. - PMC - PubMed

