Prediction of stroke severity: systematic evaluation of lesion representations
- PMID: 39394714
- PMCID: PMC11651206
- DOI: 10.1002/acn3.52215
Prediction of stroke severity: systematic evaluation of lesion representations
Abstract
Objective: To systematically evaluate which lesion-based imaging features and methods allow for the best statistical prediction of poststroke deficits across independent datasets.
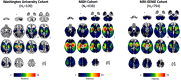
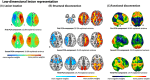
Methods: We utilized imaging and clinical data from three independent datasets of patients experiencing acute stroke (N1 = 109, N2 = 638, N3 = 794) to statistically predict acute stroke severity (NIHSS) based on lesion volume, lesion location, and structural and functional disconnection with the lesion location using normative connectomes.
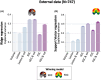
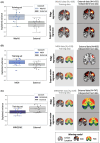
Results: We found that prediction models trained on small single-center datasets could perform well using within-dataset cross-validation, but results did not generalize to independent datasets (median R2 N1 = 0.2%). Performance across independent datasets improved using large single-center training data (R2 N2 = 15.8%) and improved further using multicenter training data (R2 N3 = 24.4%). These results were consistent across lesion attributes and prediction models. Including either structural or functional disconnection in the models outperformed prediction based on volume or location alone (P < 0.001, FDR-corrected).
Interpretation: We conclude that (1) prediction performance in independent datasets of patients with acute stroke cannot be inferred from cross-validated results within a dataset, as performance results obtained via these two methods differed consistently, (2) prediction performance can be improved by training on large and, importantly, multicenter datasets, and (3) structural and functional disconnection allow for improved prediction of acute stroke severity.
© 2024 The Author(s). Annals of Clinical and Translational Neurology published by Wiley Periodicals LLC on behalf of American Neurological Association.
Conflict of interest statement
R.W.R. has served on a DSMB for a trial sponsored by Rapid Medical and has served as site PI for studies sponsored by Penumbra and Microvention. N.S.R. has received compensation as scientific advisory consultant from Omniox, Sanofi Genzyme, and AbbVie Inc. Further authors do not have anything to disclose.
Figures


References
MeSH terms
Grants and funding
- R01MH113929/NH/NIH HHS/United States
- R25 NS065743/NS/NINDS NIH HHS/United States
- R01 NS082285/NS/NINDS NIH HHS/United States
- U19NS115388/NIH-NINDS
- Society of Vascular and Interventional Neurology
- R01MH115949/NH/NIH HHS/United States
- K23MH120510/MH/NIMH NIH HHS/United States
- R01NS086905/NIH-NINDS
- Nancy Lurie Marks Foundation
- R21 MH126271/MH/NIMH NIH HHS/United States
- R56 AG069086/AG/NIA NIH HHS/United States
- K23 MH120510/MH/NIMH NIH HHS/United States
- R25NS065743/NIH-NINDS
- R01 MH115949/MH/NIMH NIH HHS/United States
- R01 MH113929/MH/NIMH NIH HHS/United States
- Kaye Family Research Endowment
- R01NS082285/NIH-NINDS
- R01 AG060987/AG/NIA NIH HHS/United States
- R01 NS086905/NS/NINDS NIH HHS/United States
- Simons Foundation Autism Research Initiative Bridge to Independence Fellowship
- Child Neurology Foundation
- U19 NS115388/NS/NINDS NIH HHS/United States
- Heitman Foundation for Stroke
- R56AG069086/NH/NIH HHS/United States
- R01AG060987/NH/NIH HHS/United States
- R21MH126271/NH/NIH HHS/United States
- Ellison/Baszucki Foundation
LinkOut - more resources
Full Text Sources
Medical
