Comparison of the Effects of Inappropriate Meal Timing-Induced and Genetic Models of Circadian Clock Disruption on Uterine mRNA Expression Profiles
- PMID: 39395574
- PMCID: PMC11662237
- DOI: 10.1016/j.tjnut.2024.10.011
Comparison of the Effects of Inappropriate Meal Timing-Induced and Genetic Models of Circadian Clock Disruption on Uterine mRNA Expression Profiles
Abstract
Background: Accumulating evidence reveals that inappropriate meal timing contributes to the development of lifestyle-related diseases. An underlying mechanism is thought to be the disruption of the intracellular circadian clock in various tissues based on observations in both systemic and tissue-specific clock gene-deficient mice. However, whether the effects of conditional clock gene knockout are comparable to those of inappropriate meal timing remains unclear.
Objectives: This study aimed to compare the effects of a recently developed 28-h feeding cycle model with those of a core clock gene Bmal1 uterine conditional knockout (Bmal1 cKO) model on uterine mRNA expression profiles.
Methods: The models were generated by subjecting C57BL/6J mice to an 8-h/20-h feeding/fasting cycle for 2 wk and crossing Bmal1-floxed mice with PR-Cre mice. Microarray analyses were conducted using uterine samples obtained at the beginning of the dark and light periods.
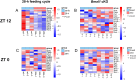
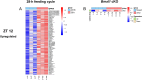
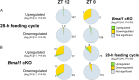
Results: The analyses identified 516 and 346, significantly 4-fold and 2-fold, up- or downregulated genes in the 28-h feeding cycle and Bmal1 cKO groups, respectively, compared with each control group. Among these genes, only 7 (1.4%) and 63 (18.2%) were significantly up- or downregulated in the other model. Moreover, most (n = 44, 62.9%) of these genes were oppositely regulated. These findings were confirmed by gene set enrichment analyses.
Conclusions: This study reveals that a 28-h feeding cycle and Bmal1 cKO differently affect gene expression profiles and highlights the need for considering this difference to assess the pathophysiology of diseases associated with inappropriate meal timing.
Keywords: circadian clock; clock genes; gene expression profiles; irregular eating habits; timing of eating; uterus.
Copyright © 2024 American Society for Nutrition. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest statement The authors report no conflicts of interest.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

