Proteomic Characterization of 1000 Human and Murine Neutrophils Freshly Isolated From Blood and Sites of Sterile Inflammation
- PMID: 39395581
- PMCID: PMC11630641
- DOI: 10.1016/j.mcpro.2024.100858
Proteomic Characterization of 1000 Human and Murine Neutrophils Freshly Isolated From Blood and Sites of Sterile Inflammation
Abstract
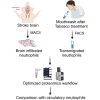
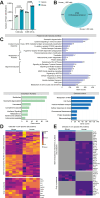
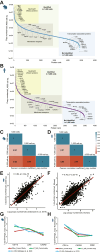
Neutrophils are indispensable for defense against pathogens. Injured tissue-infiltrated neutrophils can establish a niche of chronic inflammation and promote degeneration. Studies investigated transcriptome of single-infiltrated neutrophils which could misinterpret molecular states of these post mitotic cells. However, neutrophil proteome characterization has been challenging due to low harvests from affected tissues. Here, we present a workflow to obtain proteome of 1000 murine and human tissue-infiltrated neutrophils. We generated spectral libraries containing ∼6200 mouse and ∼5300 human proteins from circulating neutrophils. 4800 mouse and 3400 human proteins were recovered from 1000 cells with 102-108 copies/cell. Neutrophils from stroke-affected mouse brains adapted to the glucose-deprived environment with increased mitochondrial activity and ROS-production, while cells invading inflamed human oral cavities increased phagocytosis and granule release. We provide an extensive protein repository for resting human and mouse neutrophils, identify proteins lost in low input samples, thus enabling the proteomic characterization of limited tissue-infiltrated neutrophils.
Keywords: circulation; inflammation; low input proteomics; neutrophils; stroke; transmigration.
Copyright © 2024 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflicts of interests The authors declare no competing interests.
Figures





References
-
- Hedrick C.C., Malanchi I. Neutrophils in cancer: heterogeneous and multifaceted. Nat. Rev. Immunol. 2022;22:173–187. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

