Cancer-associated foam cells hamper protective T cell immunity and favor tumor progression in human colon carcinogenesis
- PMID: 39395839
- PMCID: PMC11474856
- DOI: 10.1136/jitc-2024-009720
Cancer-associated foam cells hamper protective T cell immunity and favor tumor progression in human colon carcinogenesis
Abstract
Background: Colorectal cancer (CRC) remains a significant healthcare burden worldwide, characterized by a complex interplay between obesity and chronic inflammation. While the relationship between CRC, obesity and altered lipid metabolism is not fully understood, there are evidences suggesting a link between them. In this study, we hypothesized that dysregulated lipid metabolism contributes to local accumulation of foam cells (FC) in CRC, which in turn disrupts antitumor immunosurveillance.
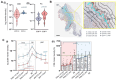
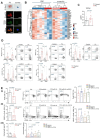
Methods: Tumor infiltrating FC and CD8+ were quantified by digital pathology in patients affected by T2-T4 CRC with any N stage undergoing radical upfront surgery (n=65) and correlated with patients' clinical outcomes. Multiparametric high-resolution flow cytometry analysis and bulk RNAseq of CRC tissue were conducted to evaluate the phenotype and transcriptomic program of immune cell infiltrate in relation to FC accumulation. The immunosuppressive effects of FC and mechanistic studies on FC-associated transforming growth factor-beta (TGF-β) and anti-PD-L1 inhibition were explored using an in-vitro human model of lipid-engulfed macrophages.
Results: FC (large CD68+ Bodipy+ macrophages) accumulated at the tumor margin in CRC samples. FChigh tumors exhibited reduced CD8+ T cells and increased regulatory T cells (Tregs). Functional transcriptional profiling depicted an immunosuppressed milieu characterized by reduced interferon gamma, memory CD8+ T cells, and activated macrophages mirrored by increased T-cell exhaustion and Treg enrichment. Furthermore, FChigh tumor phenotype was independent of standard clinical factors but correlated with high body mass index (BMI) and plasma saturated fatty acid levels. In CD8low tumors, the FChigh phenotype was associated with a 3-year disease-free survival rate of 8.6% compared with 28.7% of FClow (p=0.001). In-vitro studies demonstrated that FC significantly impact on CD8 proliferation in TFG-β dependent manner, while inhibition of TGF-β FC-related factors restored antitumor immunity.
Conclusions: FC exert immunosuppressive activity through a TGF-β-related pathway, resulting in a CD8-excluded microenvironment and identifying immunosuppressed tumors with worse prognosis in patients with primary CRC. FC association with patient BMI and dyslipidemia might explain the link of CRC with obesity, and offers novel therapeutic and preventive perspectives in this specific clinical setting.
Keywords: Colorectal Cancer; Immunosuppression; Macrophage; Tumor microenvironment - TME.
© Author(s) (or their employer(s)) 2024. Re-use permitted under CC BY-NC. No commercial re-use. See rights and permissions. Published by BMJ.
Conflict of interest statement
Competing interests: None declared.
Figures




References
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
