Entamoeba muris mitigates metabolic consequences of high-fat diet in mice
- PMID: 39396247
- PMCID: PMC11485694
- DOI: 10.1080/19490976.2024.2409210
Entamoeba muris mitigates metabolic consequences of high-fat diet in mice
Erratum in
-
Correction.Gut Microbes. 2024 Jan-Dec;16(1):2421106. doi: 10.1080/19490976.2024.2421106. Epub 2024 Oct 28. Gut Microbes. 2024. PMID: 39468836 Free PMC article. No abstract available.
Abstract
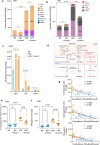
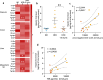
Metabolic syndrome (MetS) is a cluster of several human conditions including abdominal obesity, hypertension, dyslipidemia, and hyperglycemia, all of which are risk factors of type 2 diabetes, cardiovascular disease, and metabolic dysfunction-associated steatotic liver disease (MASLD). Dietary pattern is a well-recognized MetS risk factor, but additional changes related to the modern Western life-style may also contribute to MetS. Here we hypothesize that the disappearance of amoebas in the gut plays a role in the emergence of MetS in association with dietary changes. Four groups of C57B/6J mice fed with a high-fat diet (HFD) or a normal diet (ND) were colonized or not with Entamoeba muris, a commensal amoeba. Seventy days after inoculation, cecal microbiota, and bile acid compositions were analyzed by high-throughput sequencing of 16S rDNA and mass spectrometry, respectively. Cytokine concentrations were measured in the gut, liver, and mesenteric fat looking for low-grade inflammation. The impact of HFD on liver metabolic dysfunction was explored by Oil Red O staining, triglycerides, cholesterol concentrations, and the expression of genes involved in β-oxidation and lipogenesis. Colonization with E. muris had a beneficial impact, with a reduction in dysbiosis, lower levels of fecal secondary bile acids, and an improvement in hepatic steatosis, arguing for a protective role of commensal amoebas in MetS and more specifically HFD-associated MASLD.
Keywords: Entamoeba muris; Metabolic syndrome; amoebas; cAMP; dysbiosis; hepatic steatosis; metabolic dysfunction-associated steatotic liver disease.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures



References
-
- Muzurović E, Mikhailidis DP, Mantzoros C.. Non-alcoholic fatty liver disease, insulin resistance, metabolic syndrome and their association with vascular risk. Metabolism [Internet]. 2021. [cited 2023 Aug 17];119:154770. https://linkinghub.elsevier.com/retrieve/pii/S0026049521000706. - PubMed
-
- Cena H, Calder PC. Defining a healthy diet: evidence for the role of contemporary dietary patterns in health and disease. Nutrients [Internet]. 2020. [accessed 2023 Aug 17];12:334. https://www.mdpi.com/2072-6643/12/2/334. - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
