Fluorescence in situ Hybridization Analysis of Oligonucleotide 5S Ribosomal DNA, 45S Ribosomal DNA, and (TTTAGGG)3 Locations in Gloriosa superba L
- PMID: 39396512
- PMCID: PMC11825083
- DOI: 10.1159/000541706
Fluorescence in situ Hybridization Analysis of Oligonucleotide 5S Ribosomal DNA, 45S Ribosomal DNA, and (TTTAGGG)3 Locations in Gloriosa superba L
Abstract
Introduction: Gloriosa superba L. is a horticulturally and medicinally important plant native to Africa. However, the few cytogenetic studies of the species are mainly focused on chromosome counting and chromosome morphology-based karyotyping. Fluorescence in situ hybridization (FISH) is a powerful tool for the detection of DNA repetitive elements in a specific region of a chromosome.
Methods: Here, detailed karyotypes of G. superba were constructed by FISH using 5S and 45S rDNAs, and telomeric repeat (TTTAGGG)3 oligonucleotides.
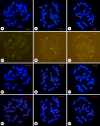
Results and conclusion: Twenty-two chromosomes were observed. Two 5S rDNA hybridization signals were detected in the proximal regions of the short arms of one pair of chromosomes, which were adjacent to the (TTTAGGG)3 terminal signals. Four 45S rDNA signals were detected near the centromere region of the short arm of the four chromosomes, but one of these was very weak and almost undetectable compared to the others. Telomeric repeat hybridization signals were distributed at the terminal region of each chromosome. The chromosomes displayed were intact, and the chromosome counts were accurate. Chromosome length ranged from 3.46 to 9.31 μm. These results will facilitate the cytogenetic mapping of other major repeats, thus contributing to an improved understanding of the G. superba genome structure and evolutionary history.
Introduction: Gloriosa superba L. is a horticulturally and medicinally important plant native to Africa. However, the few cytogenetic studies of the species are mainly focused on chromosome counting and chromosome morphology-based karyotyping. Fluorescence in situ hybridization (FISH) is a powerful tool for the detection of DNA repetitive elements in a specific region of a chromosome.
Methods: Here, detailed karyotypes of G. superba were constructed by FISH using 5S and 45S rDNAs, and telomeric repeat (TTTAGGG)3 oligonucleotides.
Results and conclusion: Twenty-two chromosomes were observed. Two 5S rDNA hybridization signals were detected in the proximal regions of the short arms of one pair of chromosomes, which were adjacent to the (TTTAGGG)3 terminal signals. Four 45S rDNA signals were detected near the centromere region of the short arm of the four chromosomes, but one of these was very weak and almost undetectable compared to the others. Telomeric repeat hybridization signals were distributed at the terminal region of each chromosome. The chromosomes displayed were intact, and the chromosome counts were accurate. Chromosome length ranged from 3.46 to 9.31 μm. These results will facilitate the cytogenetic mapping of other major repeats, thus contributing to an improved understanding of the G. superba genome structure and evolutionary history.
Keywords: Fluorescence in situ hybridization; Gloriosa; Oligonucleotide probes; Ribosomal DNA; Telomere.
© 2024 The Author(s). Published by S. Karger AG, Basel.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

References
-
- Jana S, Shekhawat GS. Critical review on medicinally potent plant species: Gloriosa superba. Fitoterapia. 2011;82(3):293–301. - PubMed
-
- Sultana SS, Dash CK, Alam SS, Hassan MA. Karyotype analysis and report on B-chromosome in Gloriosa superba L. by differential staining. Nucleus. 2018;62(1):31–8.
-
- Wu ZY, Raven PH, Hong DY, editors. Flora of China. 2nd edn.Beijing & St. Louis: Missouri Botanical Garden Press, Science Press; 2000.
-
- Mahajan R. Gloriosa superba L. : An endangered medicinal plant. Hort Flora Res Spectr. 2015;4(2):168–71.
-
- Padmapriya S, Rajamani K, Sathiyamurthy VA. Glory lily (Gloriosa superba L.)-A review. Int J Curr Pharm Rev Res. 2016;7(1):43–9.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

