Autoimmune CD4+ T cells fine-tune TCF1 expression to maintain function and survive persistent antigen exposure during diabetes
- PMID: 39396521
- PMCID: PMC11563894
- DOI: 10.1016/j.immuni.2024.09.016
Autoimmune CD4+ T cells fine-tune TCF1 expression to maintain function and survive persistent antigen exposure during diabetes
Abstract
Self-reactive T cells experience chronic antigen exposure but do not exhibit signs of exhaustion. Here, we investigated the mechanisms for sustained, functioning autoimmune CD4+ T cells despite chronic stimulation. Examination of T cell priming showed that CD4+ T cells activated in the absence of infectious signals retained TCF1 expression. At later time points and during blockade of new T cell recruitment, most islet-infiltrating autoimmune CD4+ T cells were TCF1+, although expression was reduced on a per T cell basis. The Tcf7 locus was epigenetically modified in circulating autoimmune CD4+ T cells, suggesting a pre-programmed de novo methylation of the locus in early stages of autoimmune CD4+ T cell differentiation. This mirrored the epigenetic profile of recently recruited CD4+CD62L+ T cells in the pancreas. Collectively, these data reveal a unique environment during autoimmune CD4+ T cell priming that allows T cells to fine-tune TCF1 expression and maintain long-term survival and function.
Keywords: CD4(+) T cell; TCF1; autoimmunity; exhaustion; type 1 diabetes.
Copyright © 2024 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures
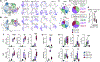

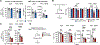




References
-
- Abdelsamed HA, Zebley CC, Nguyen H, Rutishauser RL, Fan Y, Ghoneim HE, Crawford JC, Alfei F, Alli S, Ribeiro SP, et al. (2020). Beta cell-specific CD8+ T cells maintain stem cell memory-associated epigenetic programs during type 1 diabetes. Nat Immunol 21, 578–587. 10.1038/s41590-020-0633-5. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

