DrugDAGT: a dual-attention graph transformer with contrastive learning improves drug-drug interaction prediction
- PMID: 39396972
- PMCID: PMC11472440
- DOI: 10.1186/s12915-024-02030-9
DrugDAGT: a dual-attention graph transformer with contrastive learning improves drug-drug interaction prediction
Abstract
Background: Drug-drug interactions (DDIs) can result in unexpected pharmacological outcomes, including adverse drug events, which are crucial for drug discovery. Graph neural networks have substantially advanced our ability to model molecular representations; however, the precise identification of key local structures and the capture of long-distance structural correlations for better DDI prediction and interpretation remain significant challenges.
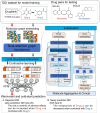
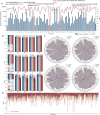
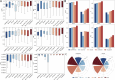
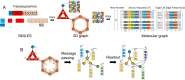
Results: Here, we present DrugDAGT, a dual-attention graph transformer framework with contrastive learning for predicting multiple DDI types. The dual-attention graph transformer incorporates attention mechanisms at both the bond and atomic levels, thereby enabling the integration of short and long-range dependencies within drug molecules to pinpoint key local structures essential for DDI discovery. Moreover, DrugDAGT further implements graph contrastive learning to maximize the similarity of representations across different views for better discrimination of molecular structures. Experiments in both warm-start and cold-start scenarios demonstrate that DrugDAGT outperforms state-of-the-art baseline models, achieving superior overall performance. Furthermore, visualization of the learned representations of drug pairs and the attention map provides interpretable insights instead of black-box results.
Conclusions: DrugDAGT provides an effective tool for accurately predicting multiple DDI types by identifying key local chemical structures, offering valuable insights for prescribing medications, and guiding drug development. All data and code of our DrugDAGT can be found at https://github.com/codejiajia/DrugDAGT .
Keywords: Attention; Drug-drug interactions; Graph transformer; Interpretation.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Sun X, Vilar S, Tatonetti NP. High-throughput methods for combinatorial drug discovery. Scienc Translational Medicine. 2013;5(205):205rv1-205rv1. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

