Immunotherapy and pan-apoptotic characterization of the tumor microenvironment in gastric cancer (STAD): a single-cell multidimensional analysis
- PMID: 39397186
- PMCID: PMC11471748
- DOI: 10.1007/s12672-024-01429-8
Immunotherapy and pan-apoptotic characterization of the tumor microenvironment in gastric cancer (STAD): a single-cell multidimensional analysis
Abstract
Background: The aim of this study was to elucidate the critical role of autophagy-related gene aggregation in gastric cancer tumor microenvironment cells and to investigate their major roles in cellular functions. In particular, the expression of these genes in tumor-associated fibroblast subtypes was scrutinized in an attempt to explain their cell-subpopulation-specific roles in cell-cell communication and regulation of cellular functions.
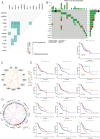
Methods: In this study, single-cell RNA sequencing data were first analyzed in multiple steps, including data preprocessing, cell clustering, and cell classification. Cell subpopulations and gene expression patterns were identified and analyzed using unsupervised non-negative matrix factorization (NMF) techniques. The dynamic expression of autophagy-related gene aggregates in various cell types was deciphered by pseudotime trajectory analysis (PTA). Intercellular communication analysis was performed using the CellChat R software package, revealing the intricate communication patterns and exchange of key signaling molecules between cell subpopulations, and SCENIC analysis was used to identify gene regulatory networks and reveal the mechanisms behind cellular heterogeneity.
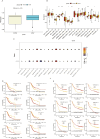
Result: Cell subpopulations associated with pan-apoptosis were identified by NMF decomposition and SCENIC analysis. Cell-cell communication analysis revealed intricate communication patterns and exchange of key signaling molecules between cell subpopulations. Dynamic expression of autophagy-related genes aggregated in the pseudotemporal trajectory of STAD was observed by PTA. In the fibroblast subtype, different ligand-receptor interactions and their key roles in immunomodulation were observed.
Conclusion: By deeply analyzing and comparing gene expression patterns within cellular subpopulations and intercellular communication, this study provides new insights into the role of pan-apoptosis-related genes in regulating immune responses and cellular functions in gastric cancer. These findings pave the way for further exploration of the role of these genes in tumorigenesis and immune regulation, as well as laying the foundation for potential therapeutic strategies.
Keywords: Immunotherapy; Molecular marker; Non-negative matrix factorization; Pan-apoptosis; Single-cell; Stomach cancer.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures








References
-
- Imounga Desroziers L, Belliardo S, Deloumeaux J, Plenet J, Bhakkan-Mambir B, Péruvien J, Chatignoux E, Macni J, Boullard M, Beaubrun-Renard M, Véronique-Baudin J, Joachim C. Burden of gastric and digestive cancers in the French Caribbean: perspectives from population-based cancer registries of Martinique, Guadeloupe and French Guiana (2007–2014). BMJ Open. 2022;12: e047167. - DOI - PMC - PubMed
-
- Wang M, Wang S, Su Q, Ma T. Effect of combining early chemotherapy with Zhipu Liujunzi Decoction under the concept of strengthening and consolidating body resistance for gastric cancer patients and nursing strategy. Contrast Media Mol Imaging. 2021;2021:2135924. 10.1155/2021/2135924. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
