LncRNA GTL2 regulates myoblast proliferation and differentiation via the PKA-CREB pathway in Duolang sheep
- PMID: 39397245
- PMCID: PMC11668951
- DOI: 10.24272/j.issn.2095-8137.2024.125
LncRNA GTL2 regulates myoblast proliferation and differentiation via the PKA-CREB pathway in Duolang sheep
Abstract
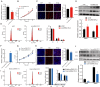
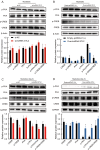
Long non-coding RNAs (lncRNAs), which are RNA molecules longer than 200 nucleotides that do not encode proteins, are implicated in a variety of biological processes, including growth and development. Despite research into the role of lncRNAs in skeletal muscle development, the regulatory mechanisms governing ovine skeletal muscle development remain unclear. In this study, we analyzed the expression profiles of lncRNAs in skeletal muscle from 90-day-old embryos (F90), 1-month-old lambs (L30), and 3-year-old adult sheep (A3Y) using RNA sequencing. In total, 4 738 lncRNAs were identified, including 997 that were differentially expressed. Short-time series expression miner analysis identified eight significant expression profiles and a subset of lncRNAs potentially involved in muscle development. Kyoto Encyclopedia of Genes and Genomes enrichment analysis revealed that the predicted target genes of these lncRNAs were primarily enriched in pathways associated with muscle development, such as the cAMP and Wnt signaling pathways. Notably, the expression of lncRNA GTL2 was found to decrease during muscle development. Moreover, GTL2 was highly expressed during the differentiation of skeletal muscle satellite cells (SCs) and was shown to modulate ovine myogenesis by affecting the phosphorylation levels of PKA and CREB. Additionally, GTL2 was found to regulate both the proliferation and differentiation of SCs via the PKA-CREB signaling pathway. Overall, this study provides a valuable resource and offers novel insights into the functional roles and regulatory mechanisms of lncRNAs in ovine skeletal muscle growth and development.
长链非编码RNA (Long non-coding RNA, lncRNAs)是一类长度超过200个核苷酸的非编码RNA,参与多种生物过程,例如生长发育。有研究报道lncRNA在骨骼肌发育中发挥重要的功能,但绵羊骨骼肌发育的调控机制尚不清楚。该研究基于RNA测序分析90日龄胚胎(F90)、1月龄羔羊(L30)和3岁成年绵羊(A3Y)骨骼肌lncRNAs的表达谱,共鉴定出4738个lncRNA,其中997个表达差异lncRNA。短时间序列基因表达(STEM)分析揭示了8个与肌肉发育相关lncRNA表达谱。KEGG富集分析显示,lncRNAs的预测靶基因主要富集于与肌肉发育相关的通路,如cAMP信号通路和Wnt信号通路等。其中,lncRNA GTL2的表达在绵羊肌肉发育过程中下降,并且lncRNA GTL2在骨骼肌卫星细胞(SCs)分化过程中高表达。此外,我们发现lncRNA GTL2通过影响PKA和CREB的蛋白磷酸化水平来调节绵羊SCs的增殖和分化。总的来说,我们的研究为探索lncRNA在羊骨骼肌发育和生长中的功能和机制提供了宝贵的数据资源和见解。.
Keywords: LncRNA; PKA-CREB pathway; Sheep; Skeletal muscle; Skeletal muscle satellite cells.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures






References
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
