Transcriptomic Biomarkers Associated With Microbiological Etiology and Disease Severity in Childhood Pneumonia
- PMID: 39397536
- PMCID: PMC11841634
- DOI: 10.1093/infdis/jiae491
Transcriptomic Biomarkers Associated With Microbiological Etiology and Disease Severity in Childhood Pneumonia
Abstract
Background: Challenges remain in discerning microbiologic etiology and disease severity in childhood pneumonia. Defining host transcriptomic profiles during illness may facilitate improved diagnostic and prognostic approaches.
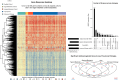
Methods: Using whole blood RNA sequencing from 222 hospitalized children with radiographic pneumonia and 45 age-matched controls, we identified differentially expressed (DE) genes that best identified children according to detected microbial pathogens (viral only vs bacterial only and typical vs atypical bacterial [with or without [±] viral co-detection]) and an ordinal measure of phenotypic severity (moderate, severe, very severe).
Results: Overall, 135 (61%) children had viral-only detections, 15 (7%) had typical bacterial detections (± viral co-detections), and 26 (12%) had atypical bacterial detections (± viral co-detections). Eleven DE genes distinguished between viral-only and bacterial-only detections. Sixteen DE genes distinguished between atypical and typical bacterial detections (± viral co-detections). Nineteen DE genes distinguished between levels of pneumonia severity, including 4 genes also identified in the viral-only versus bacterial-only model (IGHGP, PI3, CD177, RAP1GAP1) and 4 genes from the typical versus atypical bacterial model (PRSS23, IFI27, OLFM4, ABO).
Conclusions: We identified transcriptomic biomarkers associated with microbial detections and phenotypic severity in children hospitalized with pneumonia. These DE genes are promising candidates for validation and translation into diagnostic and prognostic tools.
Keywords: bacterial; pneumonia; severity scores; transcriptomics; viral.
© The Author(s) 2024. Published by Oxford University Press on behalf of Infectious Diseases Society of America.
Conflict of interest statement
Potential conflicts of interest . D. J. W. reports in-kind research support from bioMérieux for unrelated work. C. B. C. reports serving as a consultant to Pfizer, Moderna, GSK, and Vir for unrelated work. K. M. E. has received grant funding from the NIH and the CDC; has served as a consultant to Bionet and IBM; and has been a member of the data safety and monitoring board for Sanofi, X-4 Pharma, Seqirus, Moderna, Pfizer, Merck, Roche, and CEPI. O. R. reports research grant support to his institution from Janssen, Merck, NIH, and the Gates Foundation; and fees for participation in advisory boards from Sanofi Pasteur, Merck, Lilly, Adagios, and Pfizer and for lectures from Pfizer, Sanofi Pasteur, and AstraZeneca. E. J. A. receives compensation from Moderna and has consulted for Pfizer, Sanofi Pasteur, GSK, Janssen, and Medscape; serves on safety monitoring boards for Kentucky BioProcessing and Sanofi Pasteur; and serves on a data adjudication board for WCG and ACI Clinical. His institution receives funds to conduct clinical research, unrelated to this work, from MedImmune, Regeneron, PaxVax, Pfizer, GSK, Merck, Sanofi Pasteur, Janssen, and Micron, and his institution has also received funding from NIH to conduct clinical trials of COVID-19 vaccines. S. R. A. reports research grant support to her institution unrelated to this work from Pfizer, Moderna, Contrafect, and Enanta. All other authors report no potential conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Figures




References
-
- Bradley JS, Byington CL, Shah SS, et al. The management of community-acquired pneumonia in infants and children older than 3 months of age: clinical practice guidelines by the Pediatric Infectious Diseases Society and the Infectious Diseases Society of America. Clin Infect Dis 2011; 53:e25–76. - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

