Unraveling the Bivalent and Rapid Interactions Between a Multivalent RNA Recognition Motif and RNA: A Kinetic Approach
- PMID: 39397705
- PMCID: PMC11542179
- DOI: 10.1021/acs.biochem.4c00301
Unraveling the Bivalent and Rapid Interactions Between a Multivalent RNA Recognition Motif and RNA: A Kinetic Approach
Abstract
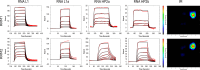
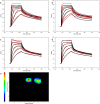
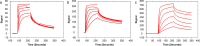
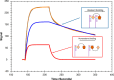
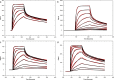
The kinetics of the interaction between Musashi-1 (MSI1) and RNA have been characterized using surface plasmon resonance biosensor analysis. Truncated variants of human MSI1 encompassing the two homologous RNA recognition motifs (RRM1 and RRM2) in tandem (aa 1-200), and the two RRMs in isolation (aa 1-103 and aa 104-200, respectively) were produced. The proteins were injected over sensor surfaces with immobilized RNA, varying in sequence and length, and with one or two RRM binding motifs. The interactions of the individual RRMs with all RNA variants were well described by a 1:1 interaction model. The interaction between the MSI1 variant encompassing both RRM motifs was bivalent and rapid for all RNA variants. Due to difficulties in fitting this complex data using standard procedures, we devised a new method to quantify the interactions. It revealed that two RRMs in tandem resulted in a significantly longer residence time than a single RRM. It also showed that RNA with double UAG binding motifs and potential hairpin structures forms less stable bivalent complexes with MSI1 than the single UAG motif containing linear RNA. Substituting the UAG binding motif with a CAG sequence resulted in a reduction of the affinity of the individual RRMs, but for MSI1, this reduction was strongly enhanced, demonstrating the importance of bivalency for specificity. This study has provided new insights into the interaction between MSI1 and RNA and an understanding of how individual domains contribute to the overall interaction. It provides an explanation for why many RNA-binding proteins contain dual RRMs.
Conflict of interest statement
The authors declare the following competing financial interest(s): Guillermo Perez-Ropero and Jos Buijs are employed by Ridgeview Instruments AB. Jos Buijs is shareholder of Ridgeview Instruments AB. Tommaso Martelli is employed by Giotto Biotech.
Figures
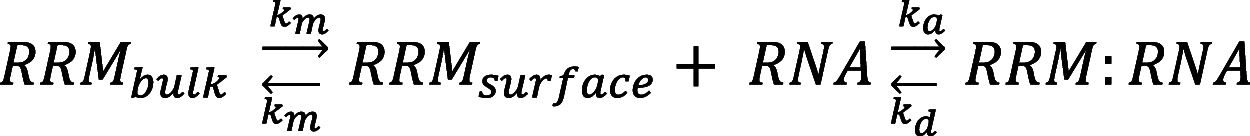



Similar articles
-
Crystal and solution structures of human oncoprotein Musashi-2 N-terminal RNA recognition motif 1.Proteins. 2020 Apr;88(4):573-583. doi: 10.1002/prot.25836. Epub 2019 Oct 29. Proteins. 2020. PMID: 31603583 Free PMC article.
-
HuD RNA recognition motifs play distinct roles in the formation of a stable complex with AU-rich RNA.Mol Cell Biol. 2000 Jul;20(13):4765-72. doi: 10.1128/MCB.20.13.4765-4772.2000. Mol Cell Biol. 2000. PMID: 10848602 Free PMC article.
-
A conserved three-nucleotide core motif defines Musashi RNA binding specificity.J Biol Chem. 2014 Dec 19;289(51):35530-41. doi: 10.1074/jbc.M114.597112. Epub 2014 Nov 3. J Biol Chem. 2014. PMID: 25368328 Free PMC article.
-
RRM-RNA recognition: NMR or crystallography…and new findings.Curr Opin Struct Biol. 2013 Feb;23(1):100-8. doi: 10.1016/j.sbi.2012.11.006. Epub 2012 Dec 14. Curr Opin Struct Biol. 2013. PMID: 23253355 Review.
-
Challenges in Therapeutically Targeting the RNA-Recognition Motif.Wiley Interdiscip Rev RNA. 2024 Nov-Dec;15(6):e1877. doi: 10.1002/wrna.1877. Wiley Interdiscip Rev RNA. 2024. PMID: 39668490 Free PMC article. Review.
Cited by
-
Deciphering the RNA recognition by Musashi-1 to design protein and RNA variants for in vitro and in vivo applications.Nucleic Acids Res. 2025 Aug 11;53(15):gkaf741. doi: 10.1093/nar/gkaf741. Nucleic Acids Res. 2025. PMID: 40795964 Free PMC article.
-
Regulatory Effects of RNA-Protein Interactions Revealed by Reporter Assays of Bacteria Grown on Solid Media.Biosensors (Basel). 2025 Mar 8;15(3):175. doi: 10.3390/bios15030175. Biosensors (Basel). 2025. PMID: 40136972 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

