Schizophrenia-associated changes in neuronal subpopulations in the human midbrain
- PMID: 39397771
- PMCID: PMC11969452
- DOI: 10.1093/brain/awae321
Schizophrenia-associated changes in neuronal subpopulations in the human midbrain
Abstract
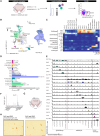
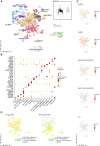
Dysfunctional GABAergic and dopaminergic neurons are thought to exist in the ventral midbrain of patients with schizophrenia, yet transcriptional changes underpinning these abnormalities have not yet been localized to specific neuronal subsets. In the ventral midbrain, control over dopaminergic activity is maintained by both excitatory (glutamate) and inhibitory (GABA) input neurons. To elucidate neuron pathology at the single-cell level, we characterized the transcriptional diversity of distinct NEUN+ populations in the human ventral midbrain and then tested for schizophrenia-associated changes in neuronal subset proportions and gene activity changes within neuronal subsets. Combining single nucleus RNA-sequencing with fluorescence-activated sorting of NEUN+ nuclei, we analysed 31 669 nuclei. Initially, we detected 18 transcriptionally distinct neuronal populations in the human ventral midbrain, including two 'mixed' populations. The presence of neuronal populations in the midbrain was orthogonally validated with immunohistochemical stainings. 'Mixed' populations contained nuclei expressing transcripts for vesicular glutamate transporter 2 (SLC17A6) and glutamate decarboxylase 2 (GAD2), but these transcripts were not typically co-expressed by the same nucleus. Upon more fine-grained subclustering of the two 'mixed' populations, 16 additional subpopulations were identified that were transcriptionally classified as excitatory or inhibitory. In the midbrains of individuals with schizophrenia, we observed potential differences in the proportions of two (sub)populations of excitatory neurons, two subpopulations of inhibitory neurons, one 'mixed' subpopulation, and one subpopulation of TH-expressing neurons. This may suggest that transcriptional changes associated with schizophrenia broadly affect excitatory, inhibitory, and dopamine neurons. We detected 99 genes differentially expressed in schizophrenia compared to controls within neuronal subpopulations identified from the two 'mixed' populations, with most (67) changes within small GABAergic neuronal subpopulations. Overall, single-nucleus transcriptomic analyses profiled a high diversity of GABAergic neurons in the human ventral midbrain, identified putative shifts in the proportion of neuronal subpopulations, and suggested dysfunction of specific GABAergic subpopulations in schizophrenia, providing directions for future research.
Keywords: midbrain; neurons; schizophrenia; single-nucleus RNA sequencing.
© The Author(s) 2024. Published by Oxford University Press on behalf of the Guarantors of Brain.
Conflict of interest statement
The authors report no competing interests.
Figures

References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

