Parallel evolution of fluconazole resistance and tolerance in Candida glabrata
- PMID: 39397866
- PMCID: PMC11466938
- DOI: 10.3389/fcimb.2024.1456907
Parallel evolution of fluconazole resistance and tolerance in Candida glabrata
Abstract
Introduction: With the growing population of immunocompromised individuals, opportunistic fungal pathogens pose a global health threat. Candida species, particularly C. albicans and non-albicans Candida species such as C. glabrata, are the most prevalent pathogenic fungi. Azoles, especially fluconazole, are widely used therapeutic options.
Objective: This study investigates how C. glabrata adapts to fluconazole, with a focus on understanding the factors regulating fluconazole tolerance and its relationship to resistance.
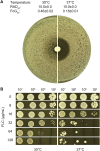
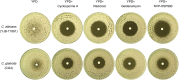
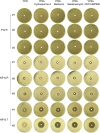
Methods: This study compared the factors regulating fluconazole tolerance between C. albicans and C. glabrata. We analyzed the impact of temperature on fluconazole tolerance, and requirement of calcineurin and Hsp90 for maintenance of fluconazole tolerance. We isolated colonies from edge, inside and outside of inhibition zone in disk diffusion assays. And we exposed C. glabrata strain to high concentrations of fluconazole and investigated the mutants for development of fluconazole resistance and tolerance.
Results: We found temperature modulated tolerance in the opposite way in C. albicans strain YJB-T1891 and C. glabrata strain CG4. Calcineurin and Hsp90 were required for maintenance of fluconazole tolerance in both species. Colonies from inside and outside of inhibition zones did not exhibited mutated phenotype, but colonies isolated from edge of inhibition zone exhibited diverse phenotype changes. Moreover, we discovered that high concentrations (16-128 μg/mL) of fluconazole induce the simultaneous but parallel development of tolerance and resistance in C. glabrata, unlike the sole development of tolerance in C. albicans.
Conclusion: This study highlights that while tolerance to fluconazole is a common response in Candida species, the specific molecular mechanisms and evolutionary pathways that lead to this response vary between species. Our findings emphasize the importance of understanding the regulation of fluconazole tolerance in different Candida species to develop effective therapeutic strategies.
Keywords: Candida glabrata; Hsp90; antifungal resistance; antifungal tolerance; calcineurin; evolution trajectory.
Copyright © 2024 Zheng, Xu, Wang, Dong and Guo.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- CLSI (2009) in 2nd ed (Wayne, PA: C.a.L.S. Institute; ).
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

