Characterization of conformational states of the homodimeric enzyme fluoroacetate dehalogenase by 19F-13C two-dimensional NMR
- PMID: 39398890
- PMCID: PMC11465415
- DOI: 10.1039/d4cb00176a
Characterization of conformational states of the homodimeric enzyme fluoroacetate dehalogenase by 19F-13C two-dimensional NMR
Abstract
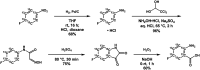
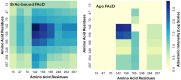
Tryptophan plays a critical role in proteins by contributing to stability, allostery, and catalysis. Using fluorine (19F) nuclear magnetic resonance (NMR), protein conformational dynamics and structure-activity relationships (SARs) can be studied via fluorotryptophan reporters. Tryptophan analogs such as 4-, 5-, 6-, or 7-fluorotryptophan can be routinely incorporated into proteins during heterologous expression by arresting endogenous tryptophan biosynthesis. Building upon the large 19F chemical shift dispersion associated with 5-fluorotryptophan, we introduce an approach to the incorporation of 13C-enriched 5-fluorotryptophan using a direct biosynthetic precursor, 5-fluoroanthranilic acid-(phenyl-13C6). The homodimeric enzyme fluoroacetate dehalogenase (FAcD), a thermophilic alpha/beta hydrolase responsible for the hydrolysis of a C-F bond in fluoroacetate, was expressed and biosynthetically labeled with (phenyl-13C6) 5-fluorotryptophan. The resulting two-dimensional 19F-13C (transverse relaxation optimized spectroscopy) TROSY heteronuclear correlation spectra provide complete resolution of all 9 tryptophan residues in the apo enzyme and FAcD saturated with the substrate analog bromoacetate. The (19F,13C) correlation spectra also reveal a multitude of minor resonances in the apo sample. The role of each tryptophan residue in allosteric communication was validated with computational rigidity transmission allostery analysis, which in this case explores the relative interprotomer communication between all possible tryptophan pairs.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
There are no conflicts to declare.
Figures
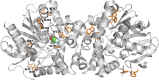


References
-
- Sharaf N. G. Gronenborn A. M. Methods Enzymol. 2015;565:67–95. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

