This is a preprint.
Loss-of-function of RNA-binding protein PRRC2B causes translational defects and congenital cardiovascular malformation
- PMID: 39398999
- PMCID: PMC11469349
- DOI: 10.1101/2024.09.26.24313895
Loss-of-function of RNA-binding protein PRRC2B causes translational defects and congenital cardiovascular malformation
Abstract
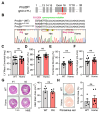
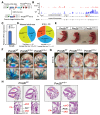
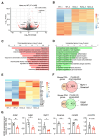
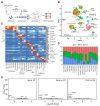
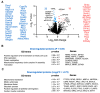
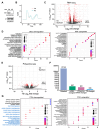
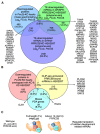
Alternative splicing generates variant forms of proteins for a given gene and accounts for functional redundancy or diversification. A novel RNA-binding protein, Pro-rich Coiled-coil Containing Protein 2B (PRRC2B), has been reported by multiple laboratories to mediate uORF-dependent and independent regulation of translation initiation required for cell cycle progression and proliferation. We identified two alternative spliced isoforms in human and mouse hearts and HEK293T cells, full-length (FL) and exon 16-excluded isoform ΔE16. A congenital heart disease-associated human mutation-mimicry knock-in of the equivalent variant in the mouse genome leads to the depletion of the full-length Prrc2b mRNA but not the alternative spliced truncated form ΔE16, does not cause any apparent structural or functional disorders. In contrast, global genetic inactivation of the PRRC2B gene in the mouse genome, nullifying both mRNA isoforms, caused patent ductus arteriosus (PDA) and neonatal lethality in mice. Bulk and single nucleus transcriptome profiling analyses of embryonic mouse hearts demonstrated a significant overall downregulation of multiple smooth muscle-specific genes in Prrc2b mutant mice resulting from reduced smooth muscle cell number. Integrated analysis of proteomic changes in Prrc2b null mouse embryonic hearts and polysome-seq and RNA-seq multi-omics analysis in human HEK293T cells uncover conserved PRRC2B-regulated target mRNAs that encode essential factors required for cardiac and vascular development. Our findings reveal the connection between alternative splicing regulation of PRRC2B, PRRC2B-mediated translational control, and congenital cardiovascular development and disorder. This study may shed light on the significance of PRRC2B in human cardiovascular disease diagnosis and treatment.
Keywords: PRRC2B; RNA-binding protein; congenital heart disease; human mutation; translation.
Conflict of interest statement
Competing interests None of the authors declares any competing interests.
Figures

References
-
- Tahmasebi S., Khoutorsky A., Mathews M.B. and Sonenberg N. (2018) Translation deregulation in human disease. Nat Rev Mol Cell Biol, 19, 791–807. - PubMed
-
- Castello A., Fischer B., Eichelbaum K., Horos R., Beckmann B.M., Strein C., Davey N.E., Humphreys D.T., Preiss T., Steinmetz L.M. et al. (2012) Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell, 149, 1393–1406. - PubMed
-
- Baltz A.G., Munschauer M., Schwanhausser B., Vasile A., Murakawa Y., Schueler M., Youngs N., Penfold-Brown D., Drew K., Milek M. et al. (2012) The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Mol Cell, 46, 674–690. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
