This is a preprint.
Resolution of SLC6A1 variable expressivity in a multi-generational family using deep clinical phenotyping and Drosophila models
- PMID: 39399018
- PMCID: PMC11469343
- DOI: 10.1101/2024.09.27.24314092
Resolution of SLC6A1 variable expressivity in a multi-generational family using deep clinical phenotyping and Drosophila models
Update in
-
Resolving SLC6A1 variable expressivity with deep clinical phenotyping and Drosophila models.HGG Adv. 2025 Oct 31;7(1):100541. doi: 10.1016/j.xhgg.2025.100541. Online ahead of print. HGG Adv. 2025. PMID: 41174879
Abstract
Purpose: Variants in SLC6A1 result in a rare neurodevelopmental disorder characterized by a variable clinical presentation of symptoms including developmental delay, epilepsy, motor dysfunction, and autism spectrum disorder. SLC6A1 haploinsufficiency has been confirmed as the predominant pathway of SLC6A1-related neurodevelopmental disorders (NDDs), however, the molecular mechanism underlying the variable clinical presentation remains unclear.
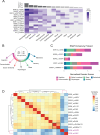
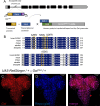
Methods: Here, through work of the Undiagnosed Diseases Network, we identify an undiagnosed individual with an inherited p.(A334S) variant of uncertain significance. To resolve this case and better understand the variable expressivity with SLC6A1, we assess the phenotypes of the proband with a cohort of cases diagnosed with SLC6A1-related NDDs. We then create an allelic series in the Drosophila melanogaster to functionally characterize case variants.
Results: We identify significant clinical overlap between the unsolved case and confirmed cases of SLC6A1-related NDDs and find a mild to severe clinical presentation associated with missense variants. We confirm phenotypes in flies expressing SLC6A1 variants consistent with a partial loss-of-function mechanism.
Conclusion: We conclude that the p.(A334S) variant is a hypomorphic allele and begin to elucidate the underlying variability in SLC6A1-related NDDs. These insights will inform clinical diagnosis, prognosis, treatment and inform therapeutic design for those living with SLC6A1-related NDDs.
Figures



References
-
- Conti F., Minelli A. & Melone M. GABA transporters in the mammalian cerebral cortex: localization, development and pathological implications. Brain Res. Brain Res. Rev. 45, 196–212 (2004). - PubMed
-
- Yuan F.-F., Gu X., Huang X., Zhong Y. & Wu J. SLC6A1 gene involvement in susceptibility to attention-deficit/hyperactivity disorder: A case-control study and gene-environment interaction. Prog. Neuropsychopharmacol. Biol. Psychiatry 77, 202–208 (2017). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
