This is a preprint.
Ancestrally diverse genome-wide association analysis highlights ancestry-specific differences in genetic regulation of plasma protein levels
- PMID: 39399032
- PMCID: PMC11469718
- DOI: 10.1101/2024.09.27.24314500
Ancestrally diverse genome-wide association analysis highlights ancestry-specific differences in genetic regulation of plasma protein levels
Abstract
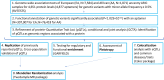
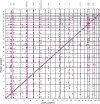
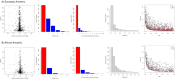
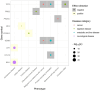
Fully characterizing the genetic architecture of circulating proteins in multi-ancestry populations provides an unprecedented opportunity to gain insights into the etiology of complex diseases. We characterized and contrasted the genetic associations of plasma proteomes in 9,455 participants of European and African (19.8%) ancestry from the Atherosclerosis Risk in Communities Study. Of 4,651 proteins, 1,408 and 2,565 proteins had protein-quantitative trait loci (pQTLs) identified in African and European ancestry respectively, and twelve unreported potentially causal protein-disease relationships were identified. Shared pQTLs across the two ancestries were detected in 1,113 aptamer-region pairs pQTLs, where 53 of them were not previously reported (all trans pQTLs). Sixteen unique protein-cardiovascular trait pairs were colocalized in both European and African ancestry with the same candidate causal variants. Our systematic cross-ancestry comparison provided a reliable set of pQTLs, highlighted the shared and distinct genetic architecture of proteome in two ancestries, and demonstrated possible biological mechanisms underlying complex diseases.
Conflict of interest statement
Conflicts of Interest Proteomic assays in ARIC were conducted free of charge as part of a data exchange agreement with SomaLogic. The authors declare no conflicts of interest related to the work submitted. R.C.H has received research grants from Denka Seiken and is a consultant (personal fees) for Denka Seiken outside the scope of the work submitted.
Figures

Similar articles
-
Protein quantitative trait locus analysis in African American and non-Hispanic White individuals.Genome Biol. 2025 Jul 10;26(1):200. doi: 10.1186/s13059-025-03671-x. Genome Biol. 2025. PMID: 40640959 Free PMC article.
-
Identification and replication of sex-dimorphic protein quantitative trait loci across multiple ancestries and their associations with diseases.Sci Rep. 2025 Aug 28;15(1):31721. doi: 10.1038/s41598-025-10031-z. Sci Rep. 2025. PMID: 40877285 Free PMC article.
-
Proteogenomic analysis integrated with electronic health records data reveals disease-associated variants in Black Americans.J Clin Invest. 2024 Sep 24;134(21):e181802. doi: 10.1172/JCI181802. J Clin Invest. 2024. PMID: 39316441 Free PMC article.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2021 Apr 19;4(4):CD011535. doi: 10.1002/14651858.CD011535.pub4. Cochrane Database Syst Rev. 2021. Update in: Cochrane Database Syst Rev. 2022 May 23;5:CD011535. doi: 10.1002/14651858.CD011535.pub5. PMID: 33871055 Free PMC article. Updated.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2017 Dec 22;12(12):CD011535. doi: 10.1002/14651858.CD011535.pub2. Cochrane Database Syst Rev. 2017. Update in: Cochrane Database Syst Rev. 2020 Jan 9;1:CD011535. doi: 10.1002/14651858.CD011535.pub3. PMID: 29271481 Free PMC article. Updated.
References
-
- Ferkingstad E, Sulem P, Atlason BA, Sveinbjornsson G, Magnusson MI, Styrmisdottir EL, et al. Large-scale integration of the plasma proteome with genetics and disease. Nat Genet 2021. December 01;53(12):1712–1721. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
