This is a preprint.
Unmasking Neuroendocrine Prostate Cancer with a Machine Learning-Driven 7-Gene Stemness Signature that Predicts Progression
- PMID: 39399052
- PMCID: PMC11469473
- DOI: 10.1101/2024.09.24.24314303
Unmasking Neuroendocrine Prostate Cancer with a Machine Learning-Driven 7-Gene Stemness Signature that Predicts Progression
Update in
-
Unmasking Neuroendocrine Prostate Cancer with a Machine Learning-Driven Seven-Gene Stemness Signature That Predicts Progression.Int J Mol Sci. 2024 Oct 22;25(21):11356. doi: 10.3390/ijms252111356. Int J Mol Sci. 2024. PMID: 39518911 Free PMC article.
Abstract
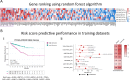
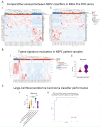
Prostate cancer (PCa) poses a significant global health challenge, particularly due to its progression into aggressive forms like neuroendocrine prostate cancer (NEPC). This study developed and validated a stemness-associated gene signature using advanced machine learning techniques, including Random Forest and Lasso regression, applied to large-scale transcriptomic datasets. The resulting 7-gene signature (KMT5C, MEN1, TYMS, IRF5, DNMT3B, CDC25B and DPP4) was validated across independent cohorts and patient-derived xenograft (PDX) models. The signature demonstrated strong prognostic value for progression-free, disease-free, relapse-free, metastasis-free, and overall survival. Importantly, the signature not only identified specific NEPC subtypes, such as large-cell neuroendocrine carcinoma, which is associated with very poor outcomes, but also predicted a poor prognosis for PCa cases that exhibit this molecular signature, even when they were not histopathologically classified as NEPC. This dual prognostic and classifier capability makes the 7-gene signature a robust tool for personalized medicine, providing a valuable resource for predicting disease progression and guiding treatment strategies in PCa management.
Keywords: Gene signature; Large Cell Neuroendocrine Carcinoma; Machine Learning; Neuroendocrine Transdifferentiation; Prognosis; Prostate Cancer; Stemness.
Conflict of interest statement
DECLARATION OF COMPETING INTEREST The authors declare no conflict of interest.
Figures




References
-
- Global Cancer Statistics 2022: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries - Bray - 2024 - CA: A Cancer Journal for Clinicians - Wiley Online Library Available online: https://acsjournals.onlinelibrary.wiley.com/doi/10.3322/caac.21834 (accessed on 18 July 2024). - DOI - PubMed
-
- Beltran H.; Rickman D.S.; Park K.; Chae S.S.; Sboner A.; MacDonald T.Y.; Wang Y.; Sheikh K.L.; Terry S.; Tagawa S.T.; et al. Molecular Characterization of Neuroendocrine Prostate Cancer and Identification of New Drug Targets. Cancer Discov. 2011, 1, 487–495, doi:10.1158/2159-8290.CD-11-0130. - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
