Integrating text mining with network models for successful target identification: in vitro validation in MASH-induced liver fibrosis
- PMID: 39399467
- PMCID: PMC11466758
- DOI: 10.3389/fphar.2024.1442752
Integrating text mining with network models for successful target identification: in vitro validation in MASH-induced liver fibrosis
Abstract
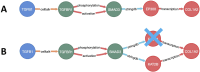
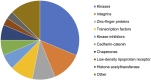
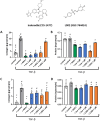
An in silico target discovery pipeline was developed by including a directional and weighted molecular disease network for metabolic dysfunction-associated steatohepatitis (MASH)-induced liver fibrosis. This approach integrates text mining, network biology, and artificial intelligence/machine learning with clinical transcriptome data for optimal translational power. At the mechanistic level, the critical components influencing disease progression were identified from the disease network using in silico knockouts. The top-ranked genes were then subjected to a target efficacy analysis, following which the top-5 candidate targets were validated in vitro. Three targets, including EP300, were confirmed for their roles in liver fibrosis. EP300 gene-silencing was found to significantly reduce collagen by 37%; compound intervention studies performed in human primary hepatic stellate cells and the hepatic stellate cell line LX-2 showed significant inhibition of collagen to the extent of 81% compared to the TGFβ-stimulated control (1 μM inobrodib in LX-2 cells). The validated in silico pipeline presents a unique approach for the identification of human-disease-mechanism-relevant drug targets. The directionality of the network ensures adherence to physiologically relevant signaling cascades, while the inclusion of clinical data boosts its translational power and ensures identification of the most relevant disease pathways. In silico knockouts thus provide crucial molecular insights for successful target identification.
Keywords: disease network; drug discovery; liver fibrosis; metabolic dysfunction-associated steatohepatitis (MASH); systems biology; target discovery; target validation; text mining.
Copyright © 2024 Venhorst, Hanemaaijer, Dulos, Caspers, Toet, Attema, de Ruiter, Kalkman, Rouhani Rankouhi, de Jong and Verschuren.
Conflict of interest statement
JV, RH, RD, MC, KT, JA, CR, GK, TRR, JJ, and LV were employed by The Netherlands Organization for Applied Scientific Research (TNO).
Figures



References
-
- Bird S., Klein E., Loper E. (2016). NLTK book. CA, USA: O’Reilly.
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

