This is a preprint.
MECHANISMS OF TECOVIRIMAT ANTIVIRAL ACTIVITY AND POXVIRUS RESISTANCE
- PMID: 39399667
- PMCID: PMC11469519
- DOI: 10.21203/rs.3.rs-5002222/v1
MECHANISMS OF TECOVIRIMAT ANTIVIRAL ACTIVITY AND POXVIRUS RESISTANCE
Update in
-
Structural insights into tecovirimat antiviral activity and poxvirus resistance.Nat Microbiol. 2025 Mar;10(3):734-748. doi: 10.1038/s41564-025-01936-6. Epub 2025 Feb 12. Nat Microbiol. 2025. PMID: 39939832 Free PMC article.
Abstract
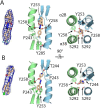
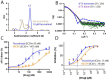
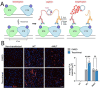
Mpox is a zoonotic disease endemic in central and west Africa. However, since 2022, human-adapted mpox virus (MPXV) strains are causing large outbreaks spreading outside these regions, leading the World Health Organization to declare public health emergency twice. Tecovirimat, the most widely used drug to treat these infections, blocks viral egress through a poorly understood mechanism. Tecovirimat-resistant strains, all with mutations in the viral phospholipase F13, pose public health concerns. Herein, we report the structure of an F13 homodimer, both alone and in complex with tecovirimat. We demonstrate that tecovirimat acts as a molecular glue, inducing the dimerization of the phospholipase. F13 escape mutations in MPXV clinical isolates are at the dimer interface and prevent drug-induced dimerization in solution and cells. These findings, which decipher tecovirimat's mode of action, will allow better monitoring of poxvirus outbreaks and pave the way for developing more potent and resilient therapeutics.
Conflict of interest statement
Competing interests: Authors declare that they have no competing interests.
Figures



References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

