AntigenBoost: enhanced mRNA-based antigen expression through rational amino acid substitution
- PMID: 39400114
- PMCID: PMC11472322
- DOI: 10.1093/bib/bbae468
AntigenBoost: enhanced mRNA-based antigen expression through rational amino acid substitution
Abstract
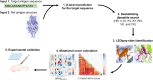
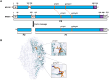
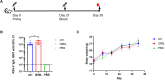
Messenger RNA (mRNA) vaccines represent a groundbreaking advancement in immunology and public health, particularly highlighted by their role in combating the COVID-19 pandemic. Optimizing mRNA-based antigen expression is a crucial focus in this emerging industry. We have developed a bioinformatics tool named AntigenBoost to address the challenge posed by destabilizing dipeptides that hinder ribosomal translation. AntigenBoost identifies these dipeptides within specific antigens and provides a range of potential amino acid substitution strategies using a two-dimensional scoring system. Through a combination of bioinformatics analysis and experimental validation, we significantly enhanced the in vitro expression of mRNA-derived Respiratory Syncytial Virus fusion glycoprotein and Influenza A Hemagglutinin antigen. Notably, a single amino acid substitution improved the immune response in mice, underscoring the effectiveness of AntigenBoost in mRNA vaccine design.
Keywords: amino acid substitution; mRNA optimization; mRNA vaccine and therapeutics; nascent peptides.
© The Author(s) 2024. Published by Oxford University Press.
Figures


Similar articles
-
A decavalent composite mRNA vaccine against both influenza and COVID-19.mBio. 2024 Sep 11;15(9):e0066824. doi: 10.1128/mbio.00668-24. Epub 2024 Aug 6. mBio. 2024. PMID: 39105586 Free PMC article.
-
A chimeric mRNA vaccine of S-RBD with HA conferring broad protection against influenza and COVID-19 variants.PLoS Pathog. 2024 Sep 20;20(9):e1012508. doi: 10.1371/journal.ppat.1012508. eCollection 2024 Sep. PLoS Pathog. 2024. PMID: 39303003 Free PMC article.
-
Advancement in the development of mRNA-based vaccines for respiratory viruses.Immunology. 2024 Nov;173(3):481-496. doi: 10.1111/imm.13844. Epub 2024 Aug 19. Immunology. 2024. PMID: 39161170 Review.
-
Molecular epidemiology and vaccine compatibility analysis of seasonal influenza A viruses in the context of COVID-19 epidemic in Wuhan, China.J Med Virol. 2024 Oct;96(10):e29858. doi: 10.1002/jmv.29858. J Med Virol. 2024. PMID: 39370830
-
The 2020 race towards SARS-CoV-2 specific vaccines.Theranostics. 2021 Jan 1;11(4):1690-1702. doi: 10.7150/thno.53691. eCollection 2021. Theranostics. 2021. PMID: 33408775 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

