Dynamic microbiome diversity shaping the adaptation of sponge holobionts in coastal waters
- PMID: 39400157
- PMCID: PMC11537060
- DOI: 10.1128/spectrum.01448-24
Dynamic microbiome diversity shaping the adaptation of sponge holobionts in coastal waters
Abstract
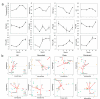
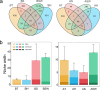
The microbial communities associated with sponges contribute to the adaptation of hosts to environments, which are essential for the trophic transformation of benthic-marine coupling. However, little is known about the symbiotic microbial community interactions and adaptative strategies of high- and low-microbial abundance (HMA and LMA) sponges, which represent two typical ecological phenotypes. Here, we compared the 1-year dynamic patterns of microbiomes with the HMA sponge Spongia officinalis and two LMA sponge species Tedania sp. and Haliclona simulans widespread on the coast of China. Symbiotic bacterial communities with the characteristic HMA-LMA dichotomy presented higher diversity and stability in S. officinalis than in Tedania sp. and H. simulans, while archaeal communities showed consistent diversity across all sponges throughout the year. Dissolved oxygen, dissolved inorganic phosphorus, dissolved organic phosphorus, and especially temperature were the major factors affecting the seasonal changes in sponge microbial communities. S. officinalis-associated microbiome had higher diversity, stronger stability, and closer interaction, which adopted a relatively isolated strategy to cope with environmental changes, while Tedania sp. and H. simulans were more susceptible and shared more bacterial Amplicon Sequence Variants (ASVs) with surrounding waters, with an open way facing the uncertainty of the environment. Meta-analysis of the microbiome in composition, diversity, and ecological function from 13 marine sponges further supported that bacterial communities associated with HMA and LMA sponges have evolved two distinct environmental adaptation strategies. We propose that the different adaptive ways of sponges responding to the environment may be responsible for their successful evolution and their competence in global ocean change.
Importance: During long-term evolution, sponge holobionts, among the oldest symbiotic relationships between microbes and metazoans, developed two distinct phenotypes with high- and low-microbial abundance (HMA and LMA). Despite sporadic studies indicating that the characteristic microbial assemblages present in HMA and LMA sponges, the adaptation strategies of symbionts responding to environments are still unclear. This deficiency limits our understanding of the selection of symbionts and the ecological functions during the evolutionary history and the adaptative assessment of HMA and LMA sponges in variable environments. Here, we explored symbiotic communities with two distinct phenotypes in a 1-year dynamic environment and combined with the meta-analysis of 13 sponges. The different strategies of symbionts in adapting to the environment were basically drawn: microbes with LMA were more acclimated to environmental changes, forming relatively loose-connected communities, while HMA developed relatively tight-connected and more similar communities beyond the divergence of species and geographical location.
Keywords: coastal zones; environmental adaptative strategy; meta-analysis of microbiome; sponge holobiont; symbiotic microbial community; temporal variation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Bang C, Dagan T, Deines P, Dubilier N, Duschl WJ, Fraune S, Hentschel U, Hirt H, Hülter N, Lachnit T, Picazo D, Pita L, Pogoreutz C, Rädecker N, Saad MM, Schmitz RA, Schulenburg H, Voolstra CR, Weiland-Bräuer N, Ziegler M, Bosch TCG. 2018. Metaorganisms in extreme environments: do microbes play a role in organismal adaptation? Zoology (Jena) 127:1–19. doi:10.1016/j.zool.2018.02.004 - DOI - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources

