Cluster-efficient pangenome graph construction with nf-core/pangenome
- PMID: 39400346
- PMCID: PMC11568064
- DOI: 10.1093/bioinformatics/btae609
Cluster-efficient pangenome graph construction with nf-core/pangenome
Abstract
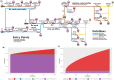
Motivation: Pangenome graphs offer a comprehensive way of capturing genomic variability across multiple genomes. However, current construction methods often introduce biases, excluding complex sequences or relying on references. The PanGenome Graph Builder (PGGB) addresses these issues. To date, though, there is no state-of-the-art pipeline allowing for easy deployment, efficient and dynamic use of available resources, and scalable usage at the same time.
Results: To overcome these limitations, we present nf-core/pangenome, a reference-unbiased approach implemented in Nextflow following nf-core's best practices. Leveraging biocontainers ensures portability and seamless deployment in High-Performance Computing (HPC) environments. Unlike PGGB, nf-core/pangenome distributes alignments across cluster nodes, enabling scalability. Demonstrating its efficiency, we constructed pangenome graphs for 1000 human chromosome 19 haplotypes and 2146 Escherichia coli sequences, achieving a two to threefold speedup compared to PGGB without increasing greenhouse gas emissions.
Availability and implementation: nf-core/pangenome is released under the MIT open-source license, available on GitHub and Zenodo, with documentation accessible at https://nf-co.re/pangenome/docs/usage.
© The Author(s) 2024. Published by Oxford University Press.
Conflict of interest statement
Author L.H. is employed by LaminLabs.
Figures
References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

