Systems analysis of miR-199a/b-5p and multiple miR-199a/b-5p targets during chondrogenesis
- PMID: 39401064
- PMCID: PMC11473111
- DOI: 10.7554/eLife.89701
Systems analysis of miR-199a/b-5p and multiple miR-199a/b-5p targets during chondrogenesis
Abstract
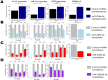
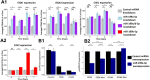
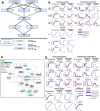
Changes in chondrocyte gene expression can contribute to the development of osteoarthritis (OA), and so recognition of the regulative processes during chondrogenesis can lead to a better understanding of OA. microRNAs (miRNAs) are key regulators of gene expression in chondrocytes/OA, and we have used a combined experimental, bioinformatic, and systems biology approach to explore the multiple miRNA-mRNA interactions that regulate chondrogenesis. A longitudinal chondrogenesis bioinformatic analysis identified paralogues miR-199a-5p and miR-199b-5p as pro-chondrogenic regulators. Experimental work in human cells demonstrated alteration of miR-199a-5p or miR-199b-5p expression led to significant inverse modulation of key chondrogenic genes and extracellular matrix production. miR-199a/b-5p targets FZD6, ITGA3 and CAV1 were identified by inhibition experiments and verified as direct targets by luciferase assay. The experimental work was used to generate and parameterise a multi-miRNA 14-day chondrogenesis kinetic model to be used as a repository for the experimental work and as a resource for further investigation of this system. This is the first multi-miRNA model of a chondrogenesis-based system, and highlights the complex relationships between regulatory miRNAs, and their target mRNAs.
Keywords: bioinformatics; chondrogenesis; computational biology; human; kinetic modelling; microRNA; mouse; osteoarthritis; systems biology.
© 2023, Patel, Barter et al.
Conflict of interest statement
KP, MB, JS, PC, CP, IC, DY, DS No competing interests declared
Figures





Update of
- doi: 10.1101/2023.03.31.535081
- doi: 10.7554/eLife.89701.1
- doi: 10.7554/eLife.89701.2
- doi: 10.7554/eLife.89701.3
References
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

