Root-specific theanine metabolism and regulation at the single-cell level in tea plants (Camellia sinensis)
- PMID: 39401074
- PMCID: PMC11473105
- DOI: 10.7554/eLife.95891
Root-specific theanine metabolism and regulation at the single-cell level in tea plants (Camellia sinensis)
Abstract
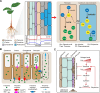
Root-synthesized secondary metabolites are critical quality-conferring compounds of foods, plant-derived medicines, and beverages. However, information at a single-cell level on root-specific secondary metabolism remains largely unexplored. L-Theanine, an important quality component of tea, is primarily synthesized in roots, from which it is then transported to new shoots of tea plant. In this study, we present a single-cell RNA sequencing (scRNA-seq)-derived map for the tea plant root, which enabled cell-type-specific analysis of glutamate and ethylamine (two precursors of theanine biosynthesis) metabolism, and theanine biosynthesis, storage, and transport. Our findings support a model in which the theanine biosynthesis pathway occurs via multicellular compartmentation and does not require high co-expression levels of transcription factors and their target genes within the same cell cluster. This study provides novel insights into theanine metabolism and regulation, at the single-cell level, and offers an example for studying root-specific secondary metabolism in other plant systems.
Keywords: Camellia sinensis; developmental biology; metabolism; plant biology; regulation; root development; scRNA-seq; theanine.
© 2024, Lin, Zhang et al.
Conflict of interest statement
SL, YZ, SZ, YW, MH, YD, JG, BZ, TY, EX, XW, WL, ZZ No competing interests declared
Figures

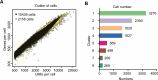






Update of
- doi: 10.1101/2024.01.16.575853
- doi: 10.7554/eLife.95891.1
- doi: 10.7554/eLife.95891.2
References
MeSH terms
Substances
Associated data
- Actions
Grants and funding
- 2022YFF1003103/National Key R&D Program of China
- 2021YFD1601101/National Key R&D Program of China
- 32072624/The National Natural Science Foundation of China
- 202103b06020024/Anhui Provincial Major Science and Technology Project
- gxyqZD2022018/Anhui Educational Committee Excellent Youth Talent Support project
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

