MSlocPRED: deep transfer learning-based identification of multi-label mRNA subcellular localization
- PMID: 39401145
- PMCID: PMC11472759
- DOI: 10.1093/bib/bbae504
MSlocPRED: deep transfer learning-based identification of multi-label mRNA subcellular localization
Abstract
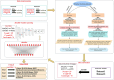
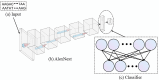
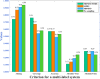
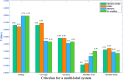
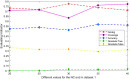
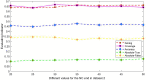
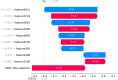
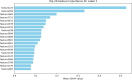
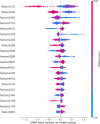
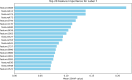
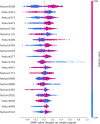
Subcellular localization of messenger ribonucleic acid (mRNA) is a universal mechanism for precise and efficient control of the translation process. Although many computational methods have been constructed by researchers for predicting mRNA subcellular localization, very few of these computational methods have been designed to predict subcellular localization with multiple localization annotations, and their generalization performance could be improved. In this study, the prediction model MSlocPRED was constructed to identify multi-label mRNA subcellular localization. First, the preprocessed Dataset 1 and Dataset 2 are transformed into the form of images. The proposed MDNDO-SMDU resampling technique is then used to balance the number of samples in each category in the training dataset. Finally, deep transfer learning was used to construct the predictive model MSlocPRED to identify subcellular localization for 16 classes (Dataset 1) and 18 classes (Dataset 2). The results of comparative tests of different resampling techniques show that the resampling technique proposed in this study is more effective in preprocessing for subcellular localization. The prediction results of the datasets constructed by intercepting different NC end (Both the 5' and 3' untranslated regions that flank the protein-coding sequence and influence mRNA function without encoding proteins themselves.) lengths show that for Dataset 1 and Dataset 2, the prediction performance is best when the NC end is intercepted by 35 nucleotides, respectively. The results of both independent testing and five-fold cross-validation comparisons with established prediction tools show that MSlocPRED is significantly better than established tools for identifying multi-label mRNA subcellular localization. Additionally, to understand how the MSlocPRED model works during the prediction process, SHapley Additive exPlanations was used to explain it. The predictive model and associated datasets are available on the following github: https://github.com/ZBYnb1/MSlocPRED/tree/main.
Keywords: deep transfer learning; interpretable analysis; sequence analysis; subcellular localization.
© The Author(s) 2024. Published by Oxford University Press.
Figures

Similar articles
-
EDCLoc: a prediction model for mRNA subcellular localization using improved focal loss to address multi-label class imbalance.BMC Genomics. 2024 Dec 27;25(1):1252. doi: 10.1186/s12864-024-11173-6. BMC Genomics. 2024. PMID: 39731012 Free PMC article.
-
mRNA-CLA: An interpretable deep learning approach for predicting mRNA subcellular localization.Methods. 2024 Jul;227:17-26. doi: 10.1016/j.ymeth.2024.04.018. Epub 2024 May 3. Methods. 2024. PMID: 38705502
-
DRpred: A Novel Deep Learning-Based Predictor for Multi-Label mRNA Subcellular Localization Prediction by Incorporating Bayesian Inferred Prior Label Relationships.Biomolecules. 2024 Aug 26;14(9):1067. doi: 10.3390/biom14091067. Biomolecules. 2024. PMID: 39334834 Free PMC article.
-
Benchmarking of Machine Learning classifiers on plasma proteomic for COVID-19 severity prediction through interpretable artificial intelligence.Artif Intell Med. 2023 Mar;137:102490. doi: 10.1016/j.artmed.2023.102490. Epub 2023 Jan 18. Artif Intell Med. 2023. PMID: 36868685 Free PMC article. Review.
-
pLoc_bal-mPlant: Predict Subcellular Localization of Plant Proteins by General PseAAC and Balancing Training Dataset.Curr Pharm Des. 2018;24(34):4013-4022. doi: 10.2174/1381612824666181119145030. Curr Pharm Des. 2018. PMID: 30451108 Review.
References
MeSH terms
Substances
Grants and funding
- PolyU152006/19E/Hong Kong Research Grants Council
- 2021YFE010178/National Key Research and Development Program of China
- JUSRP124014/Fundamental Research Funds for the Central Universities
- BK20231035/Natural Science Foundation of Jiangsu Province of China
- 62176105/National Natural Science Foundation of China
LinkOut - more resources
Full Text Sources

