Abortive infection of bat fibroblasts with SARS-CoV-2
- PMID: 39401365
- PMCID: PMC11513954
- DOI: 10.1073/pnas.2406773121
Abortive infection of bat fibroblasts with SARS-CoV-2
Abstract
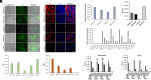
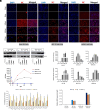
Bats are tolerant to highly pathogenic viruses such as Marburg, Ebola, and Nipah, suggesting the presence of a unique immune tolerance toward viral infection. Here, we compared severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection of human and bat (Rhinolophus ferrumequinum) pluripotent cells and fibroblasts. Since bat cells do not express an angiotensin-converting enzyme 2 (ACE2) receptor that allows virus infection, we transduced the human ACE2 (hA) receptor into the cells and found that transduced cells can be infected with SARS-CoV-2. Compared to human embryonic stem cells-hA, infected bat induced Pluripotent Stem Cells (iPSCs)-hA produced about a 100-fold lower level of infectious virus and displayed lower toxicity. In contrast, bat embryonic fibroblast-hA produced no infectious virus while being infectable and synthesizing viral RNA and proteins, suggesting abortive infection. Indeed, electron microscopy failed to detect virus-like particles in infected bat fibroblasts in contrast to bat iPSCs or human cells, consistent with the latter producing infectious viruses. This suggests that bat somatic but not pluripotent cells have an effective mechanism to control virus replication. Consistent with previous results by others, we find that bat cells have a constitutively activated innate immune system, which might limit SARS-CoV-2 infection compared to human cells.
Keywords: SARS-CoV-2; abortive infection; bat cells; innate immune system.
Conflict of interest statement
Competing interests statement:The authors declare no competing interest.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

