Lung Immune Cell Niches and the Discovery of New Cell Subtypes
- PMID: 39401416
- PMCID: PMC11615829
- DOI: 10.1002/advs.202405490
Lung Immune Cell Niches and the Discovery of New Cell Subtypes
Abstract
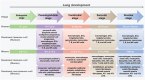
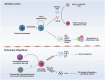
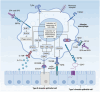
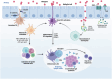
Immune cells in the lungs are important for maintaining lung function. The importance of immune cells in defending against lung diseases and infections is increasingly recognized. However, a primary knowledge gaps in current studies of lung immune cells is the understanding of their subtypes and functional heterogeneity. Increasing evidence supports the existence of novel immune cell subtypes that engage in the complex crosstalk between lung-resident immune cells, recruited immune cells, and epithelial cells. Therefore, further studies on how immune cells respond to perturbations in the pulmonary microenvironment are warranted. This review explores the processes behind the formation of the immune cell niche during lung development, and the characteristics and cell interaction modes of several major lung-resident immune cells. It indicates that distinct lung microenvironments or inflammatory niches can mediate the formation of different cell subtypes. These findings summarize and clarify paths to identify new cell subtypes that originate from resident progenitor cells and recruited peripheral cells, which are remodeled by the pulmonary microenvironment. The development of new techniques combining transcriptome analysis and location information is essential for identifying new immune cell subtypes and their relative immune niches, as well as for uncovering the molecular mechanisms of immune cell-mediated lung homeostasis.
Keywords: cell‐cell crosstalk; lung development; lung immune cells; new immune cell subtypes.
© 2024 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Hussell T., Bell T. J., Nat. Rev. Immunol. 2014, 14, 81. - PubMed
-
- a) Kalymbetova T. V., Selvakumar B., Rodríguez‐Castillo J. A., Gunjak M., Malainou C., Heindl M. R., Moiseenko A., Chao C. M., Vadász I., Mayer K., Lohmeyer J., Bellusci S., Böttcher‐Friebertshäuser E., Seeger W., Herold S., Morty R. E., J. Pathol. 2018, 245, 153. - PubMed
- b) Kierdorf K., Prinz M., Geissmann F., Perdiguero E. G., Semin. Immunol. 2015, 27, 369. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
