Evidence for a lipofibroblast-to- Cthrc1 + myofibroblast reversible switch during the development and resolution of lung fibrosis in young mice
- PMID: 39401861
- PMCID: PMC11799885
- DOI: 10.1183/13993003.00482-2023
Evidence for a lipofibroblast-to- Cthrc1 + myofibroblast reversible switch during the development and resolution of lung fibrosis in young mice
Abstract
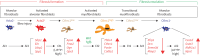
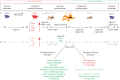
Background: Fibrosis is often associated with aberrant repair mechanisms that ultimately lead to organ failure. In the lung, idiopathic pulmonary fibrosis (IPF) is a fatal form of interstitial lung disease for which there is currently no curative therapy. From the cell biology point of view, the cell of origin and eventual fate of activated myofibroblasts (aMYFs) have taken centre stage, as these cells are believed to drive structural remodelling and lung function impairment. While aMYFs are now widely believed to originate from alveolar fibroblasts, the heterogeneity and ultimate fate of aMYFs during fibrosis resolution remain elusive. We have shown previously that aMYF dedifferentiation and acquisition of a lipofibroblast (LIF)-like phenotype represent a route of fibrosis resolution.
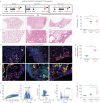
Methods: In this study, we combined genetic lineage tracing and single-cell transcriptomics in mice, and data mining of human IPF datasets to decipher the heterogeneity of aMYFs and investigate differentiation trajectories during fibrosis resolution. Furthermore, organoid cultures were utilised as a functional readout for the alveolar mesenchymal niche activity during various phases of injury and repair in mice.
Results: Our data demonstrate that aMYFs consist of four subclusters displaying unique pro-alveologenic versus pro-fibrotic profiles. Alveolar fibroblasts displaying a high LIF-like signature largely constitute both the origin and fate of aMYFs during fibrogenesis and resolution, respectively. The heterogeneity of aMYFs is conserved in humans and a significant proportion of human aMYFs displays a high LIF signature.
Conclusion: Our work identifies a subcluster of aMYFs that is potentially relevant for future management of IPF.
Copyright ©The authors 2025.
Conflict of interest statement
Conflict of interest: The authors have no potential conflicts of interest to disclose.
Figures





Comment in
-
Tracing the origins of fibrotic fibroblasts: does the name matter? Look at the genes!Eur Respir J. 2025 Feb 6;65(2):2402170. doi: 10.1183/13993003.02170-2024. Print 2025 Feb. Eur Respir J. 2025. PMID: 39915043 No abstract available.
References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
